NanoViromics: long-read sequencing of dsRNA for plant virus and viroid rapid detection
- PMID: 37415816
- PMCID: PMC10320856
- DOI: 10.3389/fmicb.2023.1192781
NanoViromics: long-read sequencing of dsRNA for plant virus and viroid rapid detection
Abstract
There is a global need for identifying viral pathogens, as well as for providing certified clean plant materials, in order to limit the spread of viral diseases. A key component of management programs for viral-like diseases is having a diagnostic tool that is quick, reliable, inexpensive, and easy to use. We have developed and validated a dsRNA-based nanopore sequencing protocol as a reliable method for detecting viruses and viroids in grapevines. We compared our method, which we term direct-cDNA sequencing from dsRNA (dsRNAcD), to direct RNA sequencing from rRNA-depleted total RNA (rdTotalRNA), and found that it provided more viral reads from infected samples. Indeed, dsRNAcD was able to detect all of the viruses and viroids detected using Illumina MiSeq sequencing (dsRNA-MiSeq). Furthermore, dsRNAcD sequencing was also able to detect low-abundance viruses that rdTotalRNA sequencing failed to detect. Additionally, rdTotalRNA sequencing resulted in a false-positive viroid identification due to the misannotation of a host-driven read. Two taxonomic classification workflows, DIAMOND & MEGAN (DIA & MEG) and Centrifuge & Recentrifuge (Cent & Rec), were also evaluated for quick and accurate read classification. Although the results from both workflows were similar, we identified pros and cons for both workflows. Our study shows that dsRNAcD sequencing and the proposed data analysis workflows are suitable for consistent detection of viruses and viroids, particularly in grapevines where mixed viral infections are common.
Keywords: Illumina Miseq sequencing; dsRNA; grapevine viruses; nanopore sequencing; plant pathology; total RNAs; viroid; virus detection.
Copyright © 2023 Javaran, Poursalavati, Lemoyne, Ste-Croix, Moffett and Fall.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
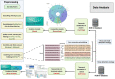
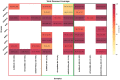
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

