Endoplasmic stress sensor Ire1 is involved in cytosolic/nuclear protein quality control in Pichia pastoris cells independent of HAC1
- PMID: 37415818
- PMCID: PMC10321714
- DOI: 10.3389/fmicb.2023.1157146
Endoplasmic stress sensor Ire1 is involved in cytosolic/nuclear protein quality control in Pichia pastoris cells independent of HAC1
Abstract
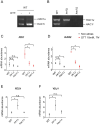
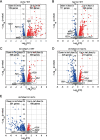
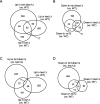
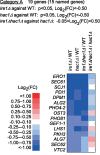
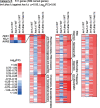
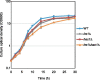
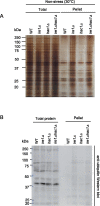
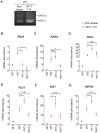
In eukaryotic species, dysfunction of the endoplasmic reticulum (ER), namely, ER stress, provokes a cytoprotective transcription program called the unfolded protein response (UPR). The UPR is triggered by transmembrane ER-stress sensors, including Ire1, which acts as an endoribonuclease to splice and mature the mRNA encoding the transcription factor Hac1 in many fungal species. Through analyses of the methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii), we revealed a previously unknown function of Ire1. In P. pastoris cells, the IRE1 knockout mutation (ire1Δ) and HAC1 knockout mutation (hac1Δ) caused only partially overlapping gene expression changes. Protein aggregation and the heat shock response (HSR) were induced in ire1Δ cells but not in hac1Δ cells even under non-stress conditions. Moreover, Ire1 was further activated upon high-temperature culturing and conferred heat stress resistance to P. pastoris cells. Our findings cumulatively demonstrate an intriguing case in which the UPR machinery controls cytosolic protein folding status and the HSR, which is known to be activated upon the accumulation of unfolded proteins in the cytosol and/or nuclei.
Keywords: Pichia pastoris; endoplasmic reticulum; stress response; unfolded protein response; yeast.
Copyright © 2023 Fauzee, Yoshida and Kimata.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts.J Fungi (Basel). 2023 Oct 5;9(10):989. doi: 10.3390/jof9100989. J Fungi (Basel). 2023. PMID: 37888245 Free PMC article. Review.
-
The unfolded protein response in Pichia pastoris without external stressing stimuli.FEMS Yeast Res. 2020 Oct 15;20(7):foaa053. doi: 10.1093/femsyr/foaa053. FEMS Yeast Res. 2020. PMID: 32926110
-
The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins.Microb Cell Fact. 2010 Jun 30;9:49. doi: 10.1186/1475-2859-9-49. Microb Cell Fact. 2010. PMID: 20591165 Free PMC article.
-
Quantitative iTRAQ LC-MS/MS proteomics reveals the cellular response to heterologous protein overexpression and the regulation of HAC1 in Pichia pastoris.J Proteomics. 2013 Oct 8;91:58-72. doi: 10.1016/j.jprot.2013.06.031. Epub 2013 Jul 11. J Proteomics. 2013. PMID: 23851310
-
The cellular response to protein misfolding in the endoplasmic reticulum.Gene Expr. 1999;7(4-6):293-300. Gene Expr. 1999. PMID: 10440230 Free PMC article. Review.
Cited by
-
Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts.J Fungi (Basel). 2023 Oct 5;9(10):989. doi: 10.3390/jof9100989. J Fungi (Basel). 2023. PMID: 37888245 Free PMC article. Review.
-
TOR and heat shock response pathways regulate peroxisome biogenesis during proteotoxic stress.bioRxiv [Preprint]. 2025 Jan 2:2024.12.31.630809. doi: 10.1101/2024.12.31.630809. bioRxiv. 2025. PMID: 40093121 Free PMC article. Preprint.
-
High-efficiency patatin expression strategies in Komagataella phaffii (Pichia pastoris): Expression cassette toolbox and regulation of protein secretion.Synth Syst Biotechnol. 2025 Jan 22;10(2):463-473. doi: 10.1016/j.synbio.2025.01.003. eCollection 2025 Jun. Synth Syst Biotechnol. 2025. PMID: 40613057 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

