The universal Sua5/TsaC family evolved different mechanisms for the synthesis of a key tRNA modification
- PMID: 37415821
- PMCID: PMC10321239
- DOI: 10.3389/fmicb.2023.1204045
The universal Sua5/TsaC family evolved different mechanisms for the synthesis of a key tRNA modification
Abstract
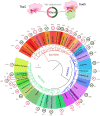
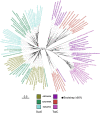
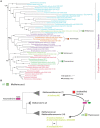
TsaC/Sua5 family of enzymes catalyzes the first step in the synthesis of N6-threonyl-carbamoyl adenosine (t6A) one of few truly ubiquitous tRNA modifications important for translation accuracy. TsaC is a single domain protein while Sua5 proteins contains a TsaC-like domain and an additional SUA5 domain of unknown function. The emergence of these two proteins and their respective mechanisms for t6A synthesis remain poorly understood. Here, we performed phylogenetic and comparative sequence and structure analysis of TsaC and Sua5 proteins. We confirm that this family is ubiquitous but the co-occurrence of both variants in the same organism is rare and unstable. We further find that obligate symbionts are the only organisms lacking sua5 or tsaC genes. The data suggest that Sua5 was the ancestral version of the enzyme while TsaC arose via loss of the SUA5 domain that occurred multiple times in course of evolution. Multiple losses of one of the two variants in combination with horizontal gene transfers along a large range of phylogenetic distances explains the present day patchy distribution of Sua5 and TsaC. The loss of the SUA5 domain triggered adaptive mutations affecting the substrate binding in TsaC proteins. Finally, we identified atypical Sua5 proteins in Archaeoglobi archaea that seem to be in the process of losing the SUA5 domain through progressive gene erosion. Together, our study uncovers the evolutionary path for emergence of these homologous isofunctional enzymes and lays the groundwork for future experimental studies on the function of TsaC/Sua5 proteins in maintaining faithful translation.
Keywords: Sua5; TsaC; enzyme; evolution; t6A; tRNA; universal proteins.
Copyright © 2023 Pichard-Kostuch, Da Cunha, Oberto, Sauguet and Basta.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Sua5 catalyzing universal t6A tRNA modification is responsible for multifaceted functions of the KEOPS complex in Cryptococcus neoformans.mSphere. 2024 Jan 30;9(1):e0055723. doi: 10.1128/msphere.00557-23. Epub 2023 Dec 12. mSphere. 2024. PMID: 38085018 Free PMC article.
-
Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.RNA. 2018 Jul;24(7):926-938. doi: 10.1261/rna.066092.118. Epub 2018 Apr 12. RNA. 2018. PMID: 29650678 Free PMC article.
-
Structure-function analysis of an ancient TsaD-TsaC-SUA5-TcdA modular enzyme reveals a prototype of tRNA t6A and ct6A synthetases.Nucleic Acids Res. 2023 Sep 8;51(16):8711-8729. doi: 10.1093/nar/gkad587. Nucleic Acids Res. 2023. PMID: 37427786 Free PMC article.
-
Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life.Int J Mol Sci. 2022 Nov 6;23(21):13600. doi: 10.3390/ijms232113600. Int J Mol Sci. 2022. PMID: 36362385 Free PMC article. Review.
-
The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification.Int J Syst Evol Microbiol. 2002 Jan;52(Pt 1):7-76. doi: 10.1099/00207713-52-1-7. Int J Syst Evol Microbiol. 2002. PMID: 11837318 Review.
Cited by
-
Sua5 catalyzing universal t6A tRNA modification is responsible for multifaceted functions of the KEOPS complex in Cryptococcus neoformans.mSphere. 2024 Jan 30;9(1):e0055723. doi: 10.1128/msphere.00557-23. Epub 2023 Dec 12. mSphere. 2024. PMID: 38085018 Free PMC article.
-
Posttranscriptional modification to the core of tRNAs modulates translational misreading errors.RNA. 2023 Dec 18;30(1):37-51. doi: 10.1261/rna.079797.123. RNA. 2023. PMID: 37907335 Free PMC article.
-
Connecting tRNA Charging and Decoding through the Axis of Nucleotide Modifications at Position 37.J Mol Biol. 2025 Aug 15;437(16):169095. doi: 10.1016/j.jmb.2025.169095. Epub 2025 Mar 18. J Mol Biol. 2025. PMID: 40113011 Review.
References
Associated data
LinkOut - more resources
Full Text Sources

