High-throughput longitudinal electrophysiology screening of mature chamber-specific hiPSC-CMs using optical mapping
- PMID: 37416454
- PMCID: PMC10320609
- DOI: 10.1016/j.isci.2023.107142
High-throughput longitudinal electrophysiology screening of mature chamber-specific hiPSC-CMs using optical mapping
Abstract
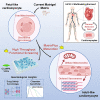
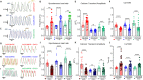
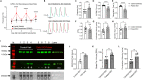
hiPSC-CMs are being considered by the Food and Drug Administration and other regulatory agencies for in vitro cardiotoxicity screening to provide human-relevant safety data. Widespread adoption of hiPSC-CMs in regulatory and academic science is limited by the immature, fetal-like phenotype of the cells. Here, to advance the maturation state of hiPSC-CMs, we developed and validated a human perinatal stem cell-derived extracellular matrix coating applied to high-throughput cell culture plates. We also present and validate a cardiac optical mapping device designed for high-throughput functional assessment of mature hiPSC-CM action potentials using voltage-sensitive dye and calcium transients using calcium-sensitive dyes or genetically encoded calcium indicators (GECI, GCaMP6). We utilize the optical mapping device to provide new biological insight into mature chamber-specific hiPSC-CMs, responsiveness to cardioactive drugs, the effect of GCaMP6 genetic variants on electrophysiological function, and the effect of daily β-receptor stimulation on hiPSC-CM monolayer function and SERCA2a expression.
Keywords: Cell biology; Medicine; Screening in health technology.
© 2023 The Author(s).
Conflict of interest statement
T.J.H. is a consultant to StemBioSys, Inc. and is a member of the Scientific Advisory Board. A.M.R., J.C., T.J.H., and T.B. have ownership stake in StemBioSys, Inc. T.B. is an employee of StemBioSys, Inc. A.A. is an employee of CAIRN Research. D.-H.K. is a scientific founder and equity holder of Curi Bio.
Figures





References
-
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
-
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

