Comparative study of multiple approaches for identifying cultivable microalgae population diversity from freshwater samples
- PMID: 37418475
- PMCID: PMC10328328
- DOI: 10.1371/journal.pone.0285913
Comparative study of multiple approaches for identifying cultivable microalgae population diversity from freshwater samples
Abstract
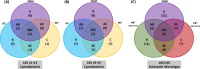
The vast diversity of microalgae imposes the challenge of identifying them through the most common and economical identification method, morphological identification, or through using the more recent molecular-level identification tools. Here we report an approach combining enrichment and metagenomic molecular techniques to enhance microalgae identification and identify microalgae diversity from environmental water samples. From this perspective, we aimed to identify the most suitable culturing media and molecular approach (using different primer sets and reference databases) for detecting microalgae diversity. Using this approach, we have analyzed three water samples collected from the River Nile on several enrichment media. A total of 37 microalgae were identified morphologically to the genus level. While sequencing the three-primer sets (16S rRNA V1-V3 and V4-V5 and 18S rRNA V4 region) and aligning them to three reference databases (GG, SILVA, and PR2), a total of 87 microalgae were identified to the genus level. The highest eukaryotic microalgae diversity was identified using the 18S rRNA V4 region and alignment to the SILVA database (43 genera). The two 16S rRNA regions sequenced added to the eukaryotic microalgae identification, 26 eukaryotic microalgae. Cyanobacteria were identified through the two sequenced 16S rRNA regions. Alignment to the SILVA database served to identify 14 cyanobacteria to the genera level, followed by Greengenes, 11 cyanobacteria genera. Our multiple-media, primer, and reference database approach revealed a high microalgae diversity that would have been overlooked if a single approach had been used over the other.
Copyright: © 2023 Badr, Fouad. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Urinary microbiome community types associated with urinary incontinence severity in women.Am J Obstet Gynecol. 2024 Mar;230(3):344.e1-344.e20. doi: 10.1016/j.ajog.2023.10.036. Epub 2023 Oct 30. Am J Obstet Gynecol. 2024. PMID: 38937257 Free PMC article.
-
Characterizing the seminal microbiota in mature rams managed on divergent planes of nutrition, and their male offspring.J Anim Sci. 2025 Jan 4;103:skaf171. doi: 10.1093/jas/skaf171. J Anim Sci. 2025. PMID: 40371922
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
Cited by
-
Metabarcoding Analysis of Microorganisms Inside Household Washing Machines in Shanghai, China.Microorganisms. 2024 Jan 13;12(1):160. doi: 10.3390/microorganisms12010160. Microorganisms. 2024. PMID: 38257987 Free PMC article.
-
Fine-Tuning of DADA2 Parameters for Multiregional Metabarcoding Analysis of 16S rRNA Genes from Activated Sludge and Comparison of Taxonomy Classification Power and Taxonomy Databases.Int J Mol Sci. 2024 Mar 20;25(6):3508. doi: 10.3390/ijms25063508. Int J Mol Sci. 2024. PMID: 38542482 Free PMC article.
References
-
- Xiao X, Sogge H, Lagesen K, Tooming-Klunderud A, Jakobsen KS, Rohrlack T. Use of High Throughput Sequencing and Light Microscopy Show Contrasting Results in a Study of Phytoplankton Occurrence in a Freshwater Environment. PLoS One [Internet]. 2014. Aug 29 [cited 2022 Apr 28];9(8):e106510. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0106510 - PMC - PubMed
-
- Kaspar U, Kriegeskorte A, Schubert T, Peters G, Rudack C, Pieper DH, et al.. The culturome of the human nose habitats reveals individual bacterial fingerprint patterns. Environ Microbiol [Internet]. 2016. Jul 1 [cited 2022 Apr 27];18(7):2130–42. Available from: https://pubmed.ncbi.nlm.nih.gov/25923378/ doi: 10.1111/1462-2920.12891 - DOI - PubMed
-
- Whelan FJ, Waddell B, Syed SA, Shekarriz S, Rabin HR, Parkins MD, et al.. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nature Microbiology 2020. 5:2 [Internet]. 2020 Jan 20 [cited 2022 Apr 27];5(2):379–90. Available from: https://www.nature.com/articles/s41564-019-0643-y doi: 10.1038/s41564-019-0643-y - DOI - PubMed
-
- Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, et al.. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nature Biotechnology 2019. 37:2 [Internet]. 2019 Feb 4 [cited 2022 Apr 27];37(2):179–85. Available from: https://www.nature.com/articles/s41587-018-0008-8 doi: 10.1038/s41587-018-0008-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

