Comparative transcriptome analysis to identify putative genes involved in carvacrol biosynthesis pathway in two species of Satureja, endemic medicinal herbs of Iran
- PMID: 37418504
- PMCID: PMC10328369
- DOI: 10.1371/journal.pone.0281351
Comparative transcriptome analysis to identify putative genes involved in carvacrol biosynthesis pathway in two species of Satureja, endemic medicinal herbs of Iran
Abstract
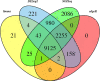
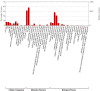
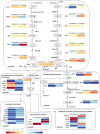
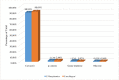
Satureja is rich in phenolic monoterpenoids, mainly carvacrol, that is of interest due to diverse biological activities including antifungal and antibacterial. However, limited information is available regarding the molecular mechanisms underlying carvacrol biosynthesis and its regulation for this wonderful medicinal herb. To identify the putative genes involved in carvacrol and other monoterpene biosynthesis pathway, we generated a reference transcriptome in two endemic Satureja species of Iran, containing different yields (Satureja khuzistanica and Satureja rechingeri). Cross-species differential expression analysis was conducted between two species of Satureja. 210 and 186 transcripts related to terpenoid backbone biosynthesis were identified for S. khuzistanica and S. rechingeri, respectively. 29 differentially expressed genes (DEGs) involved in terpenoid biosynthesis were identified, and these DEGs were significantly enriched in monoterpenoid biosynthesis, diterpenoid biosynthesis, sesquiterpenoid and triterpenoid biosynthesis, carotenoid biosynthesis and ubiquinone and other terpenoid-quinone biosynthesis pathways. Expression patterns of S. khuzistanica and S. rechingeri transcripts involved in the terpenoid biosynthetic pathway were evaluated. In addition, we identified 19 differentially expressed transcription factors (such as MYC4, bHLH, and ARF18) that may control terpenoid biosynthesis. We confirmed the altered expression levels of DEGs that encode carvacrol biosynthetic enzymes using quantitative real-time PCR (qRT-PCR). This study is the first report on de novo assembly and transcriptome data analysis in Satureja which could be useful for an understanding of the main constituents of Satureja essential oil and future research in this genus.
Copyright: © 2023 Shams et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Inhibition of the mevalonate pathway enhances carvacrol biosynthesis and DXR gene expression in shoot cultures of Satureja khuzistanica Jamzad.J Plant Physiol. 2013 Sep 1;170(13):1187-93. doi: 10.1016/j.jplph.2013.03.013. Epub 2013 Apr 20. J Plant Physiol. 2013. PMID: 23611428
-
SMRT sequencing of full-length transcriptome and gene expression analysis in two chemical types of Pogostemon cablin (Blanco) Benth.PeerJ. 2022 Feb 22;10:e12940. doi: 10.7717/peerj.12940. eCollection 2022. PeerJ. 2022. PMID: 35223208 Free PMC article.
-
Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora.BMC Genomics. 2018 Jul 24;19(1):550. doi: 10.1186/s12864-018-4941-1. BMC Genomics. 2018. PMID: 30041601 Free PMC article.
-
Recent Advances and New Insights in Genome Analysis and Transcriptomic Approaches to Reveal Enzymes Associated with the Biosynthesis of Dendrobine-Type Sesquiterpenoid Alkaloids (DTSAs) from the Last Decade.Molecules. 2024 Aug 10;29(16):3787. doi: 10.3390/molecules29163787. Molecules. 2024. PMID: 39202866 Free PMC article. Review.
-
Contemporary understanding of transcription factor regulation of terpenoid biosynthesis in plants.Planta. 2023 Nov 16;259(1):2. doi: 10.1007/s00425-023-04268-z. Planta. 2023. PMID: 37971670 Review.
Cited by
-
Comparative transcriptome analysis to identify the important mRNA and lncRNA associated with salinity tolerance in alfalfa.PeerJ. 2024 Oct 16;12:e18236. doi: 10.7717/peerj.18236. eCollection 2024. PeerJ. 2024. PMID: 39430557 Free PMC article.
References
-
- Yang W, Chen X, Li Y, Guo S, Wang Z, Yu X. Advances in pharmacological activities of terpenoids. Nat Prod Commun. 2020;15(3):1934578X20903555. 10.1177/1934578X20903555. - DOI
-
- Wang Q, Quan S, Xiao H. Towards efficient terpenoid biosynthesis: manipulating IPP and DMAPP supply. Bioresour Bioprocess. 2019;6(1): 1–3. 10.1186/s40643-019-0242-z. - DOI
-
- Croteau R, Kutchan TM, Lewis NG. Natural products (secondary metabolites). Physiol Mol Biol Plants. 2000; 24:1250–319.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

