Molecular profiling of aromatase inhibitor sensitive and resistant ER+HER2- postmenopausal breast cancers
- PMID: 37419892
- PMCID: PMC10328947
- DOI: 10.1038/s41467-023-39613-z
Molecular profiling of aromatase inhibitor sensitive and resistant ER+HER2- postmenopausal breast cancers
Abstract
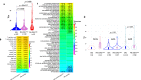
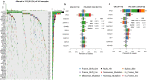
Aromatase inhibitors (AIs) reduce recurrences and mortality in postmenopausal patients with oestrogen receptor positive (ER+) breast cancer (BC), but >20% of patients will eventually relapse. Given the limited understanding of intrinsic resistance in these tumours, here we conduct a large-scale molecular analysis to identify features that impact on the response of ER + HER2- BC to AI. We compare the 15% of poorest responders (PRs, n = 177) as measured by proportional Ki67 changes after 2 weeks of neoadjuvant AI to good responders (GRs, n = 190) selected from the top 50% responders in the POETIC trial and matched for baseline Ki67 categories. In this work, low ESR1 levels are associated with poor response, high proliferation, high expression of growth factor pathways and non-luminal subtypes. PRs having high ESR1 expression have similar proportions of luminal subtypes to GRs but lower plasma estradiol levels, lower expression of estrogen response genes, higher levels of tumor infiltrating lymphocytes and immune markers, and more TP53 mutations.
© 2023. The Author(s).
Conflict of interest statement
M.D. reports consultancy for Astrazeneca, the ICR Rewards for Inventors Scheme for abiraterone. J.M.B. reports grants from Cancer Research UK, during the conduct of the study; grants from Medivation; grants and non-financial support from AstraZeneca, Merck Sharp & Dohme, Puma Biotechnology, Clovis Oncology, Pfizer, Janssen-Cilag, Novartis, and Roche, outside the submitted work. M.S.T. is a scientific advisor to Mindpeak and Sonrai Analytics, and has received honoraria recently from BMS, MSD, Roche, Sanofi and Incyte. He has received grant support from Phillips, Roche, MSD and Akoya. None of these disclosures are related to this work. J.F.R.R. has received consulting fees from, and has performed contracted research on behalf of, AstraZeneca, Bayer, Novartis and Oncimmune; has given expert testimony for AstraZeneca; and holds stock with Oncimmune. M.C.U.C. has a patent for Breast Cancer Classifier: US Patent No. 9,631,239 (Method of classifying a breast cancer instrinsic subtype) with royalties paid and receive research funding from NanoString Technologies and veracyte advisory role. M.D., E.F.S. and M.C.U.C. have patent-pending (PCT/EP2021/07368; Treatment response predictive method) for predictive CDK4/6 inhibitor sensitivity assay. All other authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

