Looping out of control: R-loops in transcription-replication conflict
- PMID: 37419963
- PMCID: PMC10771546
- DOI: 10.1007/s00412-023-00804-8
Looping out of control: R-loops in transcription-replication conflict
Abstract
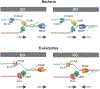
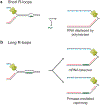
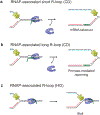
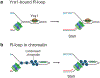
Transcription-replication conflict is a major cause of replication stress that arises when replication forks collide with the transcription machinery. Replication fork stalling at sites of transcription compromises chromosome replication fidelity and can induce DNA damage with potentially deleterious consequences for genome stability and organismal health. The block to DNA replication by the transcription machinery is complex and can involve stalled or elongating RNA polymerases, promoter-bound transcription factor complexes, or DNA topology constraints. In addition, studies over the past two decades have identified co-transcriptional R-loops as a major source for impairment of DNA replication forks at active genes. However, how R-loops impede DNA replication at the molecular level is incompletely understood. Current evidence suggests that RNA:DNA hybrids, DNA secondary structures, stalled RNA polymerases, and condensed chromatin states associated with R-loops contribute to the of fork progression. Moreover, since both R-loops and replication forks are intrinsically asymmetric structures, the outcome of R-loop-replisome collisions is influenced by collision orientation. Collectively, the data suggest that the impact of R-loops on DNA replication is highly dependent on their specific structural composition. Here, we will summarize our current understanding of the molecular basis for R-loop-induced replication fork progression defects.
Keywords: DNA replication; Genome stability; R-loops; RNA polymerase; Transcription.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




Similar articles
-
The interplay of RNA:DNA hybrid structure and G-quadruplexes determines the outcome of R-loop-replisome collisions.Elife. 2021 Sep 8;10:e72286. doi: 10.7554/eLife.72286. Elife. 2021. PMID: 34494544 Free PMC article.
-
Replisome bypass of transcription complexes and R-loops.Nucleic Acids Res. 2020 Oct 9;48(18):10353-10367. doi: 10.1093/nar/gkaa741. Nucleic Acids Res. 2020. PMID: 32926139 Free PMC article.
-
Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses.Cell. 2017 Aug 10;170(4):774-786.e19. doi: 10.1016/j.cell.2017.07.043. Cell. 2017. PMID: 28802045 Free PMC article.
-
Toxic R-loops: Cause or consequence of replication stress?DNA Repair (Amst). 2021 Nov;107:103199. doi: 10.1016/j.dnarep.2021.103199. Epub 2021 Aug 3. DNA Repair (Amst). 2021. PMID: 34399314 Review.
-
R-Loops and Its Chro-Mates: The Strange Case of Dr. Jekyll and Mr. Hyde.Int J Mol Sci. 2021 Aug 17;22(16):8850. doi: 10.3390/ijms22168850. Int J Mol Sci. 2021. PMID: 34445553 Free PMC article. Review.
Cited by
-
The hidden architects of the genome: a comprehensive review of R-loops.Mol Biol Rep. 2024 Oct 26;51(1):1095. doi: 10.1007/s11033-024-10025-6. Mol Biol Rep. 2024. PMID: 39460836 Review.
-
Functional conservation and divergence of arabidopsis VENOSA4 and human SAMHD1 in DNA repair.Heliyon. 2024 Dec 10;11(1):e41019. doi: 10.1016/j.heliyon.2024.e41019. eCollection 2025 Jan 15. Heliyon. 2024. PMID: 39801971 Free PMC article.
-
Pathological R-loops in bacteria from engineered expression of endogenous antisense RNAs whose synthesis is ordinarily terminated by Rho.Nucleic Acids Res. 2024 Nov 11;52(20):12438-12455. doi: 10.1093/nar/gkae839. Nucleic Acids Res. 2024. PMID: 39373509 Free PMC article.
-
DNA-RNA hybrids in inflammation: sources, immune response, and therapeutic implications.J Mol Med (Berl). 2025 May;103(5):511-529. doi: 10.1007/s00109-025-02533-0. Epub 2025 Mar 25. J Mol Med (Berl). 2025. PMID: 40131443 Review.
-
Minichromosome Maintenance Proteins: From DNA Replication to the DNA Damage Response.Cells. 2024 Dec 26;14(1):12. doi: 10.3390/cells14010012. Cells. 2024. PMID: 39791713 Free PMC article. Review.
References
-
- Achar YJ, Adhil M, Choudhary R, Gilbert N, Foiani M. 2020. Negative supercoil at gene boundaries modulates gene topology. Nature 577: 701–705. - PubMed
-
- Aiello U, Challal D, Wentzinger G, Lengronne A, Appanah R, Pasero P, Palancade B, Libri D. 2022. Sen1 is a key regulator of transcription-driven conflicts. Mol Cell 82: 2952–2966 e2956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

