Looping out of control: R-loops in transcription-replication conflict
- PMID: 37419963
- PMCID: PMC10771546
- DOI: 10.1007/s00412-023-00804-8
Looping out of control: R-loops in transcription-replication conflict
Abstract
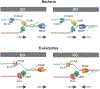
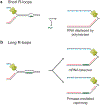
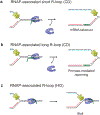
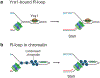
Transcription-replication conflict is a major cause of replication stress that arises when replication forks collide with the transcription machinery. Replication fork stalling at sites of transcription compromises chromosome replication fidelity and can induce DNA damage with potentially deleterious consequences for genome stability and organismal health. The block to DNA replication by the transcription machinery is complex and can involve stalled or elongating RNA polymerases, promoter-bound transcription factor complexes, or DNA topology constraints. In addition, studies over the past two decades have identified co-transcriptional R-loops as a major source for impairment of DNA replication forks at active genes. However, how R-loops impede DNA replication at the molecular level is incompletely understood. Current evidence suggests that RNA:DNA hybrids, DNA secondary structures, stalled RNA polymerases, and condensed chromatin states associated with R-loops contribute to the of fork progression. Moreover, since both R-loops and replication forks are intrinsically asymmetric structures, the outcome of R-loop-replisome collisions is influenced by collision orientation. Collectively, the data suggest that the impact of R-loops on DNA replication is highly dependent on their specific structural composition. Here, we will summarize our current understanding of the molecular basis for R-loop-induced replication fork progression defects.
Keywords: DNA replication; Genome stability; R-loops; RNA polymerase; Transcription.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




References
-
- Achar YJ, Adhil M, Choudhary R, Gilbert N, Foiani M. 2020. Negative supercoil at gene boundaries modulates gene topology. Nature 577: 701–705. - PubMed
-
- Aiello U, Challal D, Wentzinger G, Lengronne A, Appanah R, Pasero P, Palancade B, Libri D. 2022. Sen1 is a key regulator of transcription-driven conflicts. Mol Cell 82: 2952–2966 e2956. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

