Ceramide releases exosomes with a specific miRNA signature for cell differentiation
- PMID: 37419964
- PMCID: PMC10329022
- DOI: 10.1038/s41598-023-38011-1
Ceramide releases exosomes with a specific miRNA signature for cell differentiation
Abstract
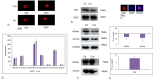
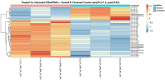
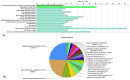
Exosomes are well established effectors of cell-cell communication. Their role on maturation of embryonic cells located in hippocampus, seat of memory, is unknown. Here we show that ceramide facilitates release of exosomes from HN9.10e cells extending information for cell differentiation to neighboring cells. We found only 38 miRNAs differentially expressed in exosomes derived from ceramide-treated cells in comparison with control cells (including 10 up-regulated and 28 down-regulated). Some overexpressed miRNAs (mmu-let-7f-1-3p, mmu-let-7a-1-3p, mmu-let-7b-3p, mmu-let-7b-5p, mmu-miR-330-3p) regulate genes encoding for protein involved in biological, homeostatic, biosynthetic and small molecule metabolic processes, embryo development and cell differentiation, all phenomena relevant for HN9.10e cell differentiation. Notably, the overexpressed mmu-let-7b-5p miRNA appears to be important for our study based on its ability to regulate thirty-five gene targets involved in many processes including sphingolipid metabolism, sphingolipid-related stimulation of cellular functions and neuronal development. Furthermore, we showed that by incubating embryonic cells with exosomes released under ceramide treatment, some cells acquired an astrocytic phenotype and others a neuronal phenotype. We anticipate our study to be a start point for innovative therapeutic strategies to regulate the release of exosomes useful to stimulate delayed brain development in the newborn and to improve the cognitive decline in neurodegenerative disorders.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sot J, Goni FM, Alonso A. Molecular associations and surface-active properties of short- and long-N-acyl chain ceramides. Biochim. Biophys. Acta. 2005;19:1711–1712. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

