A multi-omics integrative approach unravels novel genes and pathways associated with senescence escape after targeted therapy in NRAS mutant melanoma
- PMID: 37420093
- PMCID: PMC10581906
- DOI: 10.1038/s41417-023-00640-z
A multi-omics integrative approach unravels novel genes and pathways associated with senescence escape after targeted therapy in NRAS mutant melanoma
Abstract
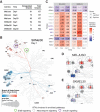
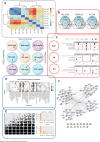
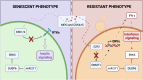
Therapy Induced Senescence (TIS) leads to sustained growth arrest of cancer cells. The associated cytostasis has been shown to be reversible and cells escaping senescence further enhance the aggressiveness of cancers. Chemicals specifically targeting senescent cells, so-called senolytics, constitute a promising avenue for improved cancer treatment in combination with targeted therapies. Understanding how cancer cells evade senescence is needed to optimise the clinical benefits of this therapeutic approach. Here we characterised the response of three different NRAS mutant melanoma cell lines to a combination of CDK4/6 and MEK inhibitors over 33 days. Transcriptomic data show that all cell lines trigger a senescence programme coupled with strong induction of interferons. Kinome profiling revealed the activation of Receptor Tyrosine Kinases (RTKs) and enriched downstream signaling of neurotrophin, ErbB and insulin pathways. Characterisation of the miRNA interactome associates miR-211-5p with resistant phenotypes. Finally, iCell-based integration of bulk and single-cell RNA-seq data identifies biological processes perturbed during senescence and predicts 90 new genes involved in its escape. Overall, our data associate insulin signaling with persistence of a senescent phenotype and suggest a new role for interferon gamma in senescence escape through the induction of EMT and the activation of ERK5 signaling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

