The salivary proteome in relation to oral mucositis in autologous hematopoietic stem cell transplantation recipients: a labelled and label-free proteomics approach
- PMID: 37420206
- PMCID: PMC10329372
- DOI: 10.1186/s12903-023-03190-w
The salivary proteome in relation to oral mucositis in autologous hematopoietic stem cell transplantation recipients: a labelled and label-free proteomics approach
Abstract
Background: Oral mucositis is a frequently seen complication in the first weeks after hematopoietic stem cell transplantation recipients which can severely affects patients quality of life. In this study, a labelled and label-free proteomics approach were used to identify differences between the salivary proteomes of autologous hematopoietic stem cell transplantation (ASCT) recipients developing ulcerative oral mucositis (ULC-OM; WHO score ≥ 2) or not (NON-OM).
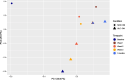
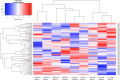
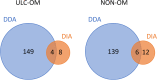
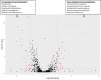
Methods: In the TMT-labelled analysis we pooled saliva samples from 5 ULC-OM patients at each of 5 timepoints: baseline, 1, 2, 3 weeks and 3 months after ASCT and compared these with pooled samples from 5 NON-OM patients. For the label-free analysis we analyzed saliva samples from 9 ULC-OM and 10 NON-OM patients at 6 different timepoints (including 12 months after ASCT) with Data-Independent Acquisition (DIA). As spectral library, all samples were grouped (ULC-OM vs NON-OM) and analyzed with Data Dependent Analysis (DDA). PCA plots and a volcano plot were generated in RStudio and differently regulated proteins were analyzed using GO analysis with g:Profiler.
Results: A different clustering of ULC-OM pools was found at baseline, weeks 2 and 3 after ASCT with TMT-labelled analysis. Using label-free analysis, week 1-3 samples clustered distinctly from the other timepoints. Unique and up-regulated proteins in the NON-OM group (DDA analysis) were involved in immune system-related processes, while those proteins in the ULC-OM group were intracellular proteins indicating cell lysis.
Conclusions: The salivary proteome in ASCT recipients has a tissue protective or tissue-damage signature, that corresponded with the absence or presence of ulcerative oral mucositis, respectively.
Trial registration: The study is registered in the national trial register (NTR5760; automatically added to the International Clinical Trial Registry Platform).
Keywords: Autologous hematopoietic stem cell transplantation; Label-free quantification; Multiple myeloma; Oral mucositis; Saliva; TMT-labelled proteomics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Blijlevens N, Schwenkglenks M, Bacon P, D'Addio A, Einsele H, Maertens J, et al. Prospective oral mucositis audit: oral mucositis in patients receiving high-dose melphalan or BEAM conditioning chemotherapy–European Blood and Marrow Transplantation Mucositis Advisory Group. J Clin Oncol. 2008;26(9):1519–1525. doi: 10.1200/JCO.2007.13.6028. - DOI - PubMed
-
- Sonis ST. Pathobiology of oral mucositis: novel insights and opportunities. J Support Oncol. 2007;5(9 Suppl 4):3–11. - PubMed
-
- Wardill HR, Sonis ST, Blijlevens NMA, Van Sebille YZA, Ciorba MA, Loeffen EAH, et al. Mucositis Study Group of the Multinational Association of Supportive Care in Cancer/International Society of Oral Oncology (MASCC/ISOO). Prediction of mucositis risk secondary to cancer therapy: a systematic review of current evidence and call to action. Support Care Cancer. 2020;28(11):5059–73. 10.1007/s00520-020-05579-7. Epub 2020 Jun 26. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

