Genomic copy number variability at the genus, species and population levels impacts in situ ecological analyses of dinoflagellates and harmful algal blooms
- PMID: 37422553
- PMCID: PMC10329664
- DOI: 10.1038/s43705-023-00274-0
Genomic copy number variability at the genus, species and population levels impacts in situ ecological analyses of dinoflagellates and harmful algal blooms
Abstract
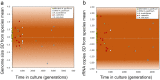
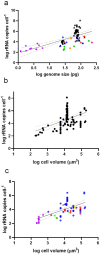
The application of meta-barcoding, qPCR, and metagenomics to aquatic eukaryotic microbial communities requires knowledge of genomic copy number variability (CNV). CNV may be particularly relevant to functional genes, impacting dosage and expression, yet little is known of the scale and role of CNV in microbial eukaryotes. Here, we quantify CNV of rRNA and a gene involved in Paralytic Shellfish Toxin (PST) synthesis (sxtA4), in 51 strains of 4 Alexandrium (Dinophyceae) species. Genomes varied up to threefold within species and ~7-fold amongst species, with the largest (A. pacificum, 130 ± 1.3 pg cell-1 /~127 Gbp) in the largest size category of any eukaryote. Genomic copy numbers (GCN) of rRNA varied by 6 orders of magnitude amongst Alexandrium (102- 108 copies cell-1) and were significantly related to genome size. Within the population CNV of rRNA was 2 orders of magnitude (105 - 107 cell-1) in 15 isolates from one population, demonstrating that quantitative data based on rRNA genes needs considerable caution in interpretation, even if validated against locally isolated strains. Despite up to 30 years in laboratory culture, rRNA CNV and genome size variability were not correlated with time in culture. Cell volume was only weakly associated with rRNA GCN (20-22% variance explained across dinoflagellates, 4% in Gonyaulacales). GCN of sxtA4 varied from 0-102 copies cell-1, was significantly related to PSTs (ng cell-1), displaying a gene dosage effect modulating PST production. Our data indicate that in dinoflagellates, a major marine eukaryotic group, low-copy functional genes are more reliable and informative targets for quantification of ecological processes than unstable rRNA genes.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Paralytic shellfish toxin content is related to genomic sxtA4 copy number in Alexandrium minutum strains.Front Microbiol. 2015 May 1;6:404. doi: 10.3389/fmicb.2015.00404. eCollection 2015. Front Microbiol. 2015. PMID: 25983733 Free PMC article.
-
From the sxtA4 Gene to Saxitoxin Production: What Controls the Variability Among Alexandrium minutum and Alexandrium pacificum Strains?Front Microbiol. 2021 Feb 24;12:613199. doi: 10.3389/fmicb.2021.613199. eCollection 2021. Front Microbiol. 2021. PMID: 33717003 Free PMC article.
-
Quantity of the dinoflagellate sxtA4 gene and cell density correlates with paralytic shellfish toxin production in Alexandrium ostenfeldii blooms.Harmful Algae. 2016 Feb;52:1-10. doi: 10.1016/j.hal.2015.10.018. Epub 2015 Dec 23. Harmful Algae. 2016. PMID: 28073466
-
Paralytic shellfish toxin biosynthesis in cyanobacteria and dinoflagellates: A molecular overview.J Proteomics. 2016 Mar 1;135:132-140. doi: 10.1016/j.jprot.2015.08.008. Epub 2015 Aug 25. J Proteomics. 2016. PMID: 26316331 Review.
-
Three decades of Canadian marine harmful algal events: Phytoplankton and phycotoxins of concern to human and ecosystem health.Harmful Algae. 2021 Feb;102:101852. doi: 10.1016/j.hal.2020.101852. Epub 2020 Jul 24. Harmful Algae. 2021. PMID: 33875179 Review.
Cited by
-
eDNA metabarcoding reveals biodiversity and depth stratification patterns of dinoflagellate assemblages within the epipelagic zone of the western Coral Sea.BMC Ecol Evol. 2024 Mar 26;24(1):38. doi: 10.1186/s12862-024-02220-7. BMC Ecol Evol. 2024. PMID: 38528460 Free PMC article.
-
Temperature-Driven Intraspecific Diversity in Paralytic Shellfish Toxin Profiles of the Dinoflagellate Alexandrium pacificum and Intragenic Variation in the Saxitoxin Biosynthetic Gene, sxtA4.Microb Ecol. 2025 Aug 8;88(1):87. doi: 10.1007/s00248-025-02586-1. Microb Ecol. 2025. PMID: 40779244 Free PMC article.
-
Potential Role of Sugars in the Hyphosphere of Arbuscular Mycorrhizal Fungi to Enhance Organic Phosphorus Mobilization.J Fungi (Basel). 2024 Mar 20;10(3):226. doi: 10.3390/jof10030226. J Fungi (Basel). 2024. PMID: 38535234 Free PMC article.
-
Quantitative metagenomics for marine prokaryotes and photosynthetic eukaryotes.ISME Commun. 2025 Jul 30;5(1):ycaf131. doi: 10.1093/ismeco/ycaf131. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40873785 Free PMC article.
-
Recovering sedimentary ancient DNA of harmful dinoflagellates accumulated over the last 9000 years off Eastern Tasmania, Australia.ISME Commun. 2024 Jul 15;4(1):ycae098. doi: 10.1093/ismeco/ycae098. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39165395 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

