Mechanisms controlling membrane recruitment and activation of the autoinhibited SHIP1 inositol 5-phosphatase
- PMID: 37423304
- PMCID: PMC10448276
- DOI: 10.1016/j.jbc.2023.105022
Mechanisms controlling membrane recruitment and activation of the autoinhibited SHIP1 inositol 5-phosphatase
Abstract
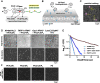
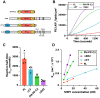
Signal transduction downstream of growth factor and immune receptor activation relies on the production of phosphatidylinositol-(3,4,5)-trisphosphate (PI(3,4,5)P3) lipids by PI3K. Regulating the strength and duration of PI3K signaling in immune cells, Src homology 2 domain-containing inositol 5-phosphatase 1 (SHIP1) controls the dephosphorylation of PI(3,4,5)P3 to generate phosphatidylinositol-(3,4)-bisphosphate. Although SHIP1 has been shown to regulate neutrophil chemotaxis, B-cell signaling, and cortical oscillations in mast cells, the role that lipid and protein interactions serve in controlling SHIP1 membrane recruitment and activity remains unclear. Using single-molecule total internal reflection fluorescence microscopy, we directly visualized membrane recruitment and activation of SHIP1 on supported lipid bilayers and the cellular plasma membrane. We find that localization of the central catalytic domain of SHIP1 is insensitive to dynamic changes in PI(3,4,5)P3 and phosphatidylinositol-(3,4)-bisphosphate both in vitro and in vivo. Very transient SHIP1 membrane interactions were detected only when membranes contained a combination of phosphatidylserine and PI(3,4,5)P3 lipids. Molecular dissection reveals that SHIP1 is autoinhibited with the N-terminal Src homology 2 domain playing a critical role in suppressing phosphatase activity. Robust SHIP1 membrane localization and relief of autoinhibition can be achieved through interactions with immunoreceptor-derived phosphopeptides presented either in solution or conjugated to a membrane. Overall, this work provides new mechanistic details concerning the dynamic interplay between lipid-binding specificity, protein-protein interactions, and the activation of autoinhibited SHIP1.
Keywords: SHIP1; Src homology 2 domain; phosphatase; phosphatidylinositol; phosphatidylinositol phosphatase; phosphatidylinositol signaling; phosphatidylinositol-3-kinase.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Update of
-
Mechanisms controlling membrane recruitment and activation of autoinhibited SHIP1.bioRxiv [Preprint]. 2023 May 1:2023.04.30.538895. doi: 10.1101/2023.04.30.538895. bioRxiv. 2023. Update in: J Biol Chem. 2023 Aug;299(8):105022. doi: 10.1016/j.jbc.2023.105022. PMID: 37205499 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

