SERPINB5 is a prognostic biomarker and promotes proliferation, metastasis and epithelial-mesenchymal transition (EMT) in lung adenocarcinoma
- PMID: 37424293
- PMCID: PMC10423661
- DOI: 10.1111/1759-7714.15013
SERPINB5 is a prognostic biomarker and promotes proliferation, metastasis and epithelial-mesenchymal transition (EMT) in lung adenocarcinoma
Abstract
Background: Serine protease inhibitors clade B serpins (SERPINBs) are the largest subclass of protease inhibitors, once thought of as a tumor suppressor gene family. However, some SERPINBs exhibit functions unrelated to the inhibition of catalytic activity.
Methods: The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Gene Set Cancer Analysis (GSCA), and cBioPortal databases were utilized to investigate SERPINBs expression, prognostic correlation, and genomic variation in 33 cancer types. We also conducted a comprehensive transcriptome analysis in multiple lung adenocarcinoma (LUAD) cohorts to reveal the molecular mechanism of SERPINB5 in LUAD. Then, qPCR and immunohistochemistry were used to verify the expression and prognostic value of SERPINB5 in LUAD patients. Furthermore, knockdown and overexpression of SERPINB5 in LUAD cell lines were performed to evaluate cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT).
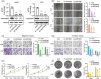
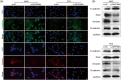
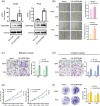
Results: The expression of SERPINB5 was upregulated and demethylated in LUAD, and its abnormally high expression was significantly correlated with poor overall survival (OS). In addition, the expression of SERPINB5 was analyzed to determine its prognostic value in LUAD and confirmed that SERPINB5 was an independent predictor of LUAD in TCGA and GEO cohorts and qPCR validation with 106 clinical samples. At last, A knockdown of SERPINB5 in LUAD cells reduced proliferation, migration, and EMT. Proliferation, migration, and invasion are promoted by the overexpression of SERPINB5.
Conclusion: Therefore, SERPINB5 has shown potential as a prognostic biomarker for LUAD, and it may become a potential therapeutic target for lung adenocarcinoma.
Keywords: EMT; SERPINB5; biomarker; lung adenocarcinoma; metastasis.
© 2023 The Authors. Thoracic Cancer published by China Lung Oncology Group and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
SGO2 as a Prognostic Biomarker Correlated with Cell Proliferation, Migration, Invasion, and Epithelial-Mesenchymal Transition in Lung Adenocarcinoma.Front Biosci (Landmark Ed). 2024 Aug 30;29(9):314. doi: 10.31083/j.fbl2909314. Front Biosci (Landmark Ed). 2024. PMID: 39344317
-
Cystatin SN promotes epithelial-mesenchymal transition and serves as a prognostic biomarker in lung adenocarcinoma.BMC Cancer. 2022 May 30;22(1):589. doi: 10.1186/s12885-022-09685-z. BMC Cancer. 2022. PMID: 35637432 Free PMC article.
-
JAM2 is a prognostic biomarker and inhibits proliferation, metastasis and epithelial-mesenchymal transition in lung adenocarcinoma.J Gene Med. 2024 Feb;26(2):e3679. doi: 10.1002/jgm.3679. J Gene Med. 2024. PMID: 38404047
-
LINC01117 inhibits invasion and migration of lung adenocarcinoma through influencing EMT process.PLoS One. 2023 Jun 29;18(6):e0287926. doi: 10.1371/journal.pone.0287926. eCollection 2023. PLoS One. 2023. PMID: 37384755 Free PMC article.
-
The paradoxical role of SERPINB5 in gastrointestinal cancers: oncogene or tumor suppressor?Mol Biol Rep. 2025 Feb 3;52(1):188. doi: 10.1007/s11033-025-10293-w. Mol Biol Rep. 2025. PMID: 39899168 Review.
Cited by
-
Machine-learning developed an iron, copper, and sulfur-metabolism associated signature predicts lung adenocarcinoma prognosis and therapy response.Respir Res. 2024 May 14;25(1):206. doi: 10.1186/s12931-024-02839-6. Respir Res. 2024. PMID: 38745285 Free PMC article.
-
Computational Analyses Identified Three Diagnostic Biomarkers Associated With Programmed Cell Death for Lung Adenocarcinoma.Hum Mutat. 2025 Aug 17;2025:1743829. doi: 10.1155/humu/1743829. eCollection 2025. Hum Mutat. 2025. PMID: 40860289 Free PMC article.
-
Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients.Cancers (Basel). 2024 Mar 10;16(6):1112. doi: 10.3390/cancers16061112. Cancers (Basel). 2024. PMID: 38539447 Free PMC article.
-
Construction of a lung adenocarcinoma prognostic model based on KEAP1/NRF2/HO‑1 mutation‑mediated upregulated genes and bioinformatic analysis.Oncol Lett. 2025 Jan 23;29(3):155. doi: 10.3892/ol.2025.14902. eCollection 2025 Mar. Oncol Lett. 2025. PMID: 39911153 Free PMC article.
-
Tumor-derived apoptotic extracellular vesicle-mediated intercellular communication promotes metastasis and stemness of lung adenocarcinoma.Bioact Mater. 2024 Mar 6;36:238-255. doi: 10.1016/j.bioactmat.2024.02.026. eCollection 2024 Jun. Bioact Mater. 2024. PMID: 38481566 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446–54. - PubMed
-
- Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. - PubMed
-
- Castellanos E, Feld E, Horn L. Driven by mutations: the predictive value of mutation subtype in EGFR‐mutated non‐small cell lung cancer. J Thorac Oncol. 2017;12:612–23. - PubMed
-
- Sun X, Li K, Zhao R, Sun Y, Xu J, Peng ZY, et al. Lung cancer pathogenesis and poor response to therapy were dependent on driver oncogenic mutations. Life Sci. 2021;265:118797. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

