Decoding the fibromelanosis locus complex chromosomal rearrangement of black-bone chicken: genetic differentiation, selective sweeps and protein-coding changes in Kadaknath chicken
- PMID: 37424723
- PMCID: PMC10325862
- DOI: 10.3389/fgene.2023.1180658
Decoding the fibromelanosis locus complex chromosomal rearrangement of black-bone chicken: genetic differentiation, selective sweeps and protein-coding changes in Kadaknath chicken
Abstract
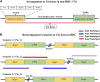
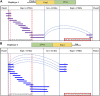
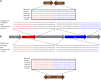
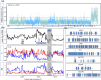
Black-bone chicken (BBC) meat is popular for its distinctive taste and texture. A complex chromosomal rearrangement at the fibromelanosis (Fm) locus on the 20th chromosome results in increased endothelin-3 (EDN3) gene expression and is responsible for melanin hyperpigmentation in BBC. We use public long-read sequencing data of the Silkie breed to resolve high-confidence haplotypes at the Fm locus spanning both Dup1 and Dup2 regions and establish that the Fm_2 scenario is correct of the three possible scenarios of the complex chromosomal rearrangement. The relationship between Chinese and Korean BBC breeds with Kadaknath native to India is underexplored. Our data from whole-genome re-sequencing establish that all BBC breeds, including Kadaknath, share the complex chromosomal rearrangement junctions at the fibromelanosis (Fm) locus. We also identify two Fm locus proximal regions (∼70 Kb and ∼300 Kb) with signatures of selection unique to Kadaknath. These regions harbor several genes with protein-coding changes, with the bactericidal/permeability-increasing-protein-like gene having two Kadaknath-specific changes within protein domains. Our results indicate that protein-coding changes in the bactericidal/permeability-increasing-protein-like gene hitchhiked with the Fm locus in Kadaknath due to close physical linkage. Identifying this Fm locus proximal selective sweep sheds light on the genetic distinctiveness of Kadaknath compared to other BBC.
Keywords: Fm locus; Kadaknath; black-bone chicken; fibromelanosis; genetic linkage.
Copyright © 2023 Shinde, Sharma and Vijay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Genetics of hyperpigmentation associated with the Fibromelanosis gene (Fm) and analysis of growth and meat quality traits in crosses of native Indian Kadaknath chickens and non-indigenous breeds.Br Poult Sci. 2011 Dec;52(6):675-85. doi: 10.1080/00071668.2011.635637. Br Poult Sci. 2011. PMID: 22221233
-
The origin and evolution of fibromelanosis in domesticated chickens: Genomic comparison of Indonesian Cemani and Chinese Silkie breeds.PLoS One. 2017 Apr 5;12(4):e0173147. doi: 10.1371/journal.pone.0173147. eCollection 2017. PLoS One. 2017. PMID: 28379963 Free PMC article.
-
A complex genomic rearrangement involving the endothelin 3 locus causes dermal hyperpigmentation in the chicken.PLoS Genet. 2011 Dec;7(12):e1002412. doi: 10.1371/journal.pgen.1002412. Epub 2011 Dec 22. PLoS Genet. 2011. PMID: 22216010 Free PMC article.
-
"Unveiling the genetic symphony: Diversity and expression of chicken IFITM genes in Aseel and Kadaknath breeds".Heliyon. 2024 Sep 10;10(18):e37729. doi: 10.1016/j.heliyon.2024.e37729. eCollection 2024 Sep 30. Heliyon. 2024. PMID: 39315180 Free PMC article.
-
An interpretive review of selective sweep studies in Bos taurus cattle populations: identification of unique and shared selection signals across breeds.Front Genet. 2015 May 13;6:167. doi: 10.3389/fgene.2015.00167. eCollection 2015. Front Genet. 2015. PMID: 26029239 Free PMC article. Review.
Cited by
-
Characterization of the Coding Sequence of the MC1R (Melanocortin 1 Receptor) Gene of Ayam Cemani Black Chickens.Animals (Basel). 2024 Aug 29;14(17):2507. doi: 10.3390/ani14172507. Animals (Basel). 2024. PMID: 39272291 Free PMC article.
-
Common Ancestry of the Id Locus: Chromosomal Rearrangement and Polygenic Possibilities.J Mol Evol. 2025 Feb;93(1):163-180. doi: 10.1007/s00239-025-10233-z. Epub 2025 Jan 17. J Mol Evol. 2025. PMID: 39821315
-
Effect of a Unique Dwarfism on Growth, Production, and Reproduction Performance of the Nicobari Chicken Breed.J Poult Sci. 2025 Mar 10;62:2025012. doi: 10.2141/jpsa.2025012. eCollection 2025. J Poult Sci. 2025. PMID: 40065752 Free PMC article.
-
Frontiers and emerging topics in a century of Silkie chicken research: insights, challenges, and opportunities.Poult Sci. 2025 May;104(5):105030. doi: 10.1016/j.psj.2025.105030. Epub 2025 Mar 12. Poult Sci. 2025. PMID: 40101517 Free PMC article. Review.
-
Reply to: The genomic structure of complex chromosomal rearrangement at the Fm locus in black-bone Silkie chicken.Commun Biol. 2025 Apr 1;8(1):536. doi: 10.1038/s42003-025-07826-1. Commun Biol. 2025. PMID: 40169757 Free PMC article. No abstract available.
References
LinkOut - more resources
Full Text Sources

