Culturable fungi from urban soils in China II, with the description of 18 novel species in Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes and Sordariomycetes)
- PMID: 37425100
- PMCID: PMC10326621
- DOI: 10.3897/mycokeys.98.102816
Culturable fungi from urban soils in China II, with the description of 18 novel species in Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes and Sordariomycetes)
Abstract
As China's urbanisation continues to advance, more people are choosing to live in cities. However, this trend has a significant impact on the natural ecosystem. For instance, the accumulation of keratin-rich substrates in urban habitats has led to an increase in keratinophilic microbes. Despite this, there is still a limited amount of research on the prevalence of keratinophilic fungi in urban areas. Fortunately, our group has conducted in-depth investigations into this topic since 2015. Through our research, we have discovered a significant amount of keratinophilic fungi in soil samples collected from various urban areas in China. In this study, we have identified and characterised 18 new species through the integration of morphological and phylogenetic analyses. These findings reveal the presence of numerous unexplored fungal taxa in urban habitats, emphasising the need for further taxonomic research in urban China.
Keywords: 18 new taxa; Fungal taxonomy; keratinophilic fungi; morphological characters; phylogeny.
Zhi-Yuan Zhang, Xin Li, Wan-Hao Chen, Jian-Dong Liang, Yan-Feng Han.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




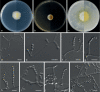





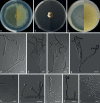





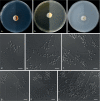






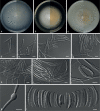
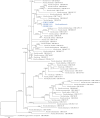
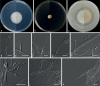



Similar articles
-
Culturable Fungi from Urban Soils in China I: Description of 10 New Taxa.Microbiol Spectr. 2021 Oct 31;9(2):e0086721. doi: 10.1128/Spectrum.00867-21. Epub 2021 Oct 6. Microbiol Spectr. 2021. PMID: 34612666 Free PMC article.
-
Diversity of Onygenalean Fungi in Keratin-Rich Habitats of Maharashtra (India) and Description of Three Novel Taxa.Mycopathologia. 2020 Feb;185(1):67-85. doi: 10.1007/s11046-019-00346-7. Epub 2019 Jun 11. Mycopathologia. 2020. PMID: 31187339
-
Lignicolous freshwater fungi in Yunnan Province, China: an overview.Mycology. 2022 Apr 3;13(2):119-132. doi: 10.1080/21501203.2022.2058638. eCollection 2022. Mycology. 2022. PMID: 35711328 Free PMC article.
-
A Review of the Fungi That Degrade Plastic.J Fungi (Basel). 2022 Jul 25;8(8):772. doi: 10.3390/jof8080772. J Fungi (Basel). 2022. PMID: 35893140 Free PMC article. Review.
-
Distribution of Keratinophilic Fungi in Soil Across Tunisia: A Descriptive Study and Review of the Literature.Mycopathologia. 2015 Aug;180(1-2):61-8. doi: 10.1007/s11046-015-9870-9. Epub 2015 Feb 18. Mycopathologia. 2015. PMID: 25690159 Review.
Cited by
-
Expanding the Toolbox for Genetic Manipulation in Pseudogymnoascus: RNAi-Mediated Silencing and CRISPR/Cas9-Mediated Disruption of a Polyketide Synthase Gene Involved in Red Pigment Production in P. verrucosus.J Fungi (Basel). 2024 Feb 16;10(2):157. doi: 10.3390/jof10020157. J Fungi (Basel). 2024. PMID: 38392828 Free PMC article.
-
Bionectriaceae: a poorly known family of hypocrealean fungi with major commercial potential.Stud Mycol. 2025 Jun;111:115-198. doi: 10.3114/sim.2025.111.04. Epub 2025 Apr 17. Stud Mycol. 2025. PMID: 40371418 Free PMC article.
-
Two new acremonium-like species, Paragibellulopsissinensis sp. nov. and Phialoparvumsinense sp. nov. (Sordariomycetes, Ascomycota) from China.MycoKeys. 2025 Jun 6;118:207-224. doi: 10.3897/mycokeys.118.155316. eCollection 2025. MycoKeys. 2025. PMID: 40521222 Free PMC article.
-
Phylogenomics, taxonomy and morphological characters of the Microdochiaceae (Xylariales, Sordariomycetes).MycoKeys. 2024 Jul 3;106:303-325. doi: 10.3897/mycokeys.106.127355. eCollection 2024. MycoKeys. 2024. PMID: 38993357 Free PMC article.
-
Three new Pseudogymnoascus species (Pseudeurotiaceae, Thelebolales) described from Antarctic soils.IMA Fungus. 2025 Mar 21;16:e142219. doi: 10.3897/imafungus.16.e142219. eCollection 2025. IMA Fungus. 2025. PMID: 40162003 Free PMC article.
References
-
- Antrop M. (2004) Landscape change and the urbanization process in Europe. Landscape and Urban Planning 67(1–4): 9–26. 10.1016/S0169-2046(03)00026-4 - DOI
-
- Auerswald B. (1869) Synopsis pyrenomycetum europaeorum. In: Gonnerman W, Rabenhorst GL (Eds) Mycologia Europaea.
-
- Benjamin CR. (1955) Ascocarps of Aspergillus and Penicillium. Mycologia 47(5): 669–687. 10.1080/00275514.1955.12024485 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
