This is a preprint.
Wolbachia -mediated resistance to Zika virus infection in Aedes aegypti is dominated by diverse transcriptional regulation and weak evolutionary pressures
- PMID: 37425681
- PMCID: PMC10327090
- DOI: 10.1101/2023.06.26.546271
Wolbachia -mediated resistance to Zika virus infection in Aedes aegypti is dominated by diverse transcriptional regulation and weak evolutionary pressures
Update in
-
Wolbachia-mediated resistance to Zika virus infection in Aedes aegypti is dominated by diverse transcriptional regulation and weak evolutionary pressures.PLoS Negl Trop Dis. 2023 Oct 2;17(10):e0011674. doi: 10.1371/journal.pntd.0011674. eCollection 2023 Oct. PLoS Negl Trop Dis. 2023. PMID: 37782672 Free PMC article.
Abstract
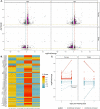
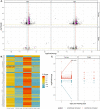
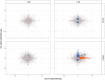
A promising candidate for arbovirus control and prevention relies on replacing arbovirus-susceptible Aedes aegypti populations with mosquitoes that have been colonized by the intracellular bacterium Wolbachia and thus have a reduced capacity to transmit arboviruses. This reduced capacity to transmit arboviruses is mediated through a phenomenon referred to as pathogen blocking. Pathogen blocking has primarily been proposed as a tool to control dengue virus (DENV) transmission, however it works against a range of viruses, including Zika virus (ZIKV). Despite years of research, the molecular mechanisms underlying pathogen blocking still need to be better understood. Here, we used RNA-seq to characterize mosquito gene transcription dynamics in Ae. aegypti infected with the w Mel strain of Wolbachia that are being released by the World Mosquito Program in Medellín, Colombia. Comparative analyses using ZIKV-infected, uninfected tissues, and mosquitoes without Wolbachia revealed that the influence of w Mel on mosquito gene transcription is multifactorial. Importantly, because Wolbachia limits, but does not completely prevent, replication of ZIKV and other viruses in coinfected mosquitoes, there is a possibility that these viruses could evolve resistance to pathogen blocking. Therefore, to understand the influence of Wolbachia on within-host ZIKV evolution, we characterized the genetic diversity of molecularly barcoded ZIKV virus populations replicating in Wolbachia -infected mosquitoes and found that within-host ZIKV evolution was subject to weak purifying selection and, unexpectedly, loose anatomical bottlenecks in the presence and absence of Wolbachia . Together, these findings suggest that there is no clear transcriptional profile associated with Wolbachia -mediated ZIKV restriction, and that there is no evidence for ZIKV escape from this restriction in our system.
Author summary: When Wolbachia bacteria infect Aedes aegypti mosquitoes, they dramatically reduce the mosquitoes' susceptibility to infection with a range of arthropod-borne viruses, including Zika virus (ZIKV). Although this pathogen-blocking effect has been widely recognized, its mechanisms remain unclear. Furthermore, because Wolbachia limits, but does not completely prevent, replication of ZIKV and other viruses in coinfected mosquitoes, there is a possibility that these viruses could evolve resistance to Wolbachia -mediated blocking. Here, we use host transcriptomics and viral genome sequencing to examine the mechanisms of ZIKV pathogen blocking by Wolbachia and viral evolutionary dynamics in Ae. aegypti mosquitoes. We find complex transcriptome patterns that do not suggest a single clear mechanism for pathogen blocking. We also find no evidence that Wolbachia exerts detectable selective pressures on ZIKV in coinfected mosquitoes. Together our data suggest that it may be difficult for ZIKV to evolve Wolbachia resistance, perhaps due to the complexity of the pathogen blockade mechanism.
Figures





References
-
- Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011. Aug;476(7361):450–3. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
