This is a preprint.
CaBLAM! A high-contrast bioluminescent Ca2+ indicator derived from an engineered Oplophorus gracilirostris luciferase
- PMID: 37425712
- PMCID: PMC10327125
- DOI: 10.1101/2023.06.25.546478
CaBLAM! A high-contrast bioluminescent Ca2+ indicator derived from an engineered Oplophorus gracilirostris luciferase
Abstract
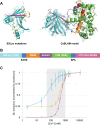
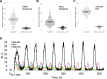
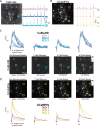
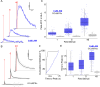
Ca2+ plays many critical roles in cell physiology and biochemistry, leading researchers to develop a number of fluorescent small molecule dyes and genetically encodable probes that optically report changes in Ca2+ concentrations in living cells. Though such fluorescence-based genetically encoded Ca2+ indicators (GECIs) have become a mainstay of modern Ca2+ sensing and imaging, bioluminescence-based GECIs-probes that generate light through oxidation of a small-molecule by a luciferase or photoprotein-have several distinct advantages over their fluorescent counterparts. Bioluminescent tags do not photobleach, do not suffer from nonspecific autofluorescent background, and do not lead to phototoxicity since they do not require the extremely bright extrinsic excitation light typically required for fluorescence imaging, especially with 2-photon microscopy. Current BL GECIs perform poorly relative to fluorescent GECIs, producing small changes in bioluminescence intensity due to high baseline signal at resting Ca2+ concentrations and suboptimal Ca2+ affinities. Here, we describe the development of a new bioluminescent GECI, "CaBLAM," which displays much higher contrast (dynamic range) than previously described bioluminescent GECIs and has a Ca2+ affinity suitable for capturing physiological changes in cytosolic Ca2+ concentration. Derived from a new variant of Oplophorus gracilirostris luciferase with superior in vitro properties and a highly favorable scaffold for insertion of sensor domains, CaBLAM allows for single-cell and subcellular resolution imaging of Ca2+ dynamics at high frame rates in cultured neurons and in vivo. CaBLAM marks a significant milestone in the GECI timeline, enabling Ca2+ recordings with high spatial and temporal resolution without perturbing cells with intense excitation light.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Genetically Encoded Fluorescence/Bioluminescence Bimodal Indicators for Ca2+ Imaging.ACS Sens. 2019 Jul 26;4(7):1825-1834. doi: 10.1021/acssensors.9b00531. Epub 2019 Jul 5. ACS Sens. 2019. PMID: 31276380
-
Bioluminescent Sensors for Ca++ Flux Imaging and the Introduction of a New Intensity-Based Ca++ Sensor.Front Bioeng Biotechnol. 2021 Oct 27;9:773353. doi: 10.3389/fbioe.2021.773353. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34778237 Free PMC article.
-
Bioluminescent Low-Affinity Ca2+ Indicator for ER with Multicolor Calcium Imaging in Single Living Cells.ACS Chem Biol. 2018 Jul 20;13(7):1862-1871. doi: 10.1021/acschembio.7b01014. Epub 2018 Mar 9. ACS Chem Biol. 2018. PMID: 29494125
-
Use of aequorin-based indicators for monitoring Ca2+ in acidic organelles.Biochim Biophys Acta Mol Cell Res. 2023 Aug;1870(6):119481. doi: 10.1016/j.bbamcr.2023.119481. Epub 2023 May 2. Biochim Biophys Acta Mol Cell Res. 2023. PMID: 37142127 Review.
-
Putting a finishing touch on GECIs.Front Mol Neurosci. 2014 Nov 18;7:88. doi: 10.3389/fnmol.2014.00088. eCollection 2014. Front Mol Neurosci. 2014. PMID: 25477779 Free PMC article. Review.
References
-
- Shen Y., Nasu Y., Shkolnikov I., Kim A. & Campbell R. E. Engineering genetically encoded fluorescent indicators for imaging of neuronal activity: progress and prospects. Neurosci Res 152, 3–14 (2020). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
