This is a preprint.
Multiple pathways for glucose phosphate transport and utilization support growth of Cryptosporidium parvum
- PMID: 37425855
- PMCID: PMC10327089
- DOI: 10.1101/2023.06.27.546703
Multiple pathways for glucose phosphate transport and utilization support growth of Cryptosporidium parvum
Update in
-
Multiple pathways for glucose phosphate transport and utilization support growth of Cryptosporidium parvum.Nat Commun. 2024 Jan 9;15(1):380. doi: 10.1038/s41467-024-44696-3. Nat Commun. 2024. PMID: 38191884 Free PMC article.
Abstract
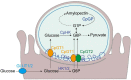
Cryptosporidium parvum is an obligate intracellular parasite with a highly reduced mitochondrion that lacks the TCA cycle and the ability to generate ATP, making the parasite reliant on glycolysis. Genetic ablation experiments demonstrated that neither of the two putative glucose transporters CpGT1 and CpGT2 were essential for growth. Surprisingly, hexokinase was also dispensable for parasite growth while the downstream enzyme aldolase was required, suggesting the parasite has an alternative way of obtaining phosphorylated hexose. Complementation studies in E. coli support a role for direct transport of glucose-6-phosphate from the host cell by the parasite transporters CpGT1 and CpGT2, thus bypassing a requirement for hexokinase. Additionally, the parasite obtains phosphorylated glucose from amylopectin stores that are released by the action of the essential enzyme glycogen phosphorylase. Collectively, these findings reveal that C. parvum relies on multiple pathways to obtain phosphorylated glucose both for glycolysis and to restore carbohydrate reserves.
Keywords: energy utilization; glucose phosphate transport; glucose transport; glycogen; glycolysis.
Conflict of interest statement
Competing Interests: The authors have no competing interests to declare.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
