This is a preprint.
Target-directed microRNA degradation regulates developmental microRNA expression and embryonic growth in mammals
- PMID: 37425885
- PMCID: PMC10327180
- DOI: 10.1101/2023.06.26.546601
Target-directed microRNA degradation regulates developmental microRNA expression and embryonic growth in mammals
Update in
-
Target-directed microRNA degradation regulates developmental microRNA expression and embryonic growth in mammals.Genes Dev. 2023 Jul 1;37(13-14):661-674. doi: 10.1101/gad.350906.123. Epub 2023 Aug 8. Genes Dev. 2023. PMID: 37553261 Free PMC article.
Abstract
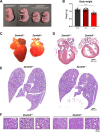
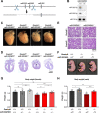
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression that play critical roles in development and disease. Target-directed miRNA degradation (TDMD), a pathway in which miRNAs that bind to specialized targets with extensive complementarity are rapidly decayed, has emerged as a potent mechanism of controlling miRNA levels. Nevertheless, the biological role and scope of miRNA regulation by TDMD in mammals remains poorly understood. To address these questions, we generated mice with constitutive or conditional deletion of Zswim8 , which encodes an essential TDMD factor. Loss of Zswim8 resulted in developmental defects in heart and lung, growth restriction, and perinatal lethality. Small RNA sequencing of embryonic tissues revealed widespread miRNA regulation by TDMD and greatly expanded the known catalog of miRNAs regulated by this pathway. These experiments also uncovered novel features of TDMD-regulated miRNAs, including their enrichment in co-transcribed clusters and examples in which TDMD underlies 'arm switching', a phenomenon wherein the dominant strand of a miRNA precursor changes in different tissues or conditions. Importantly, deletion of two miRNAs, miR-322 and miR-503, rescued growth of Zswim8 null embryos, directly implicating the TDMD pathway as a regulator of mammalian body size. These data illuminate the broad landscape and developmental role of TDMD in mammals.
Conflict of interest statement
Competing interest statement
J.T.M is a scientific advisor for Ribometrix, Inc. and owns equity in Orbital Therapeutics, Inc. The other authors declare no competing interests.
Figures




References
-
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreno Gutierrez H, Perlas E, Perez-Rico YA, Tocchini-Valentini GP, Enright AJ, Norton WHJ et al. 2018. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat Struct Mol Biol 25: 244–251. doi:10.1038/s41594-018-0032-x - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
