A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
- PMID: 37426617
- PMCID: PMC10329280
- DOI: 10.1515/biol-2022-0635
A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut
Abstract
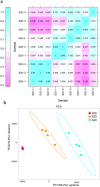
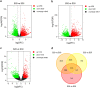
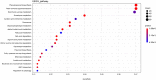
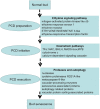
Previous studies suggest that the senescence and death of the replaceable bud of the Chinese chestnut cultivar (cv.) "Tima Zhenzhu" involves programmed cell death (PCD). However, the molecular network regulating replaceable bud PCD is poorly characterized. Here, we performed transcriptomic profiling on the chestnut cv. "Tima Zhenzhu" replaceable bud before (S20), during (S25), and after (S30) PCD to unravel the molecular mechanism underlying the PCD process. A total of 5,779, 9,867, and 2,674 differentially expressed genes (DEGs) were discovered upon comparison of S20 vs S25, S20 vs S30, and S25 vs S30, respectively. Approximately 6,137 DEGs common to at least two comparisons were selected for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to interrogate the main corresponding biological functions and pathways. GO analysis showed that these common DEGs could be divided into three functional categories, including 15 cellular components, 14 molecular functions, and 19 biological processes. KEGG analysis found that "plant hormone signal transduction" included 93 DEGs. Overall, 441 DEGs were identified as related to the process of PCD. Most of these were found to be genes associated with ethylene signaling, as well as the initiation and execution of various PCD processes.
Keywords: Chinese chestnut; programmed cell death; replaceable bud; transcriptomics.
© 2023 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: Authors state no conflict of interest.
Figures





Similar articles
-
Programmed cell death is responsible for replaceable bud senescence in chestnut (Castanea mollissima BL.).Plant Cell Rep. 2012 Sep;31(9):1603-10. doi: 10.1007/s00299-012-1274-4. Epub 2012 Jun 3. Plant Cell Rep. 2012. PMID: 22660903
-
Transcriptomic profiling identifies differentially expressed genes associated with programmed cell death of nucellar cells in Ginkgo biloba L.BMC Plant Biol. 2019 Feb 28;19(1):91. doi: 10.1186/s12870-019-1671-8. BMC Plant Biol. 2019. PMID: 30819114 Free PMC article.
-
Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape.BMC Plant Biol. 2017 Jan 19;17(1):18. doi: 10.1186/s12870-016-0960-8. BMC Plant Biol. 2017. PMID: 28103799 Free PMC article.
-
RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus).Gene. 2015 Jan 25;555(2):362-76. doi: 10.1016/j.gene.2014.11.032. Epub 2014 Nov 15. Gene. 2015. PMID: 25447903
-
Transcriptome Analysis of Litsea cubeba Floral Buds Reveals the Role of Hormones and Transcription Factors in the Differentiation Process.G3 (Bethesda). 2018 Mar 28;8(4):1103-1114. doi: 10.1534/g3.117.300481. G3 (Bethesda). 2018. PMID: 29487185 Free PMC article.
Cited by
-
Development of pollinated and unpollinated ovules in Ginkgo biloba: unravelling the role of pollen in ovule tissue maturation.J Exp Bot. 2024 Jun 7;75(11):3351-3367. doi: 10.1093/jxb/erae102. J Exp Bot. 2024. PMID: 38459807 Free PMC article.
References
-
- Wang G, Zhang Z, Kong D, Liu Q, Zhao G. Programmed cell death is responsible for replaceable bud senescence in chestnut (Castanea mollissima BL.). Plant Cell Rep. 2012;31:1603–10. - PubMed
-
- Liu Q, Kong D, Wang G. A new chestnut variety ‘Tima Zhenzhu’. Acta Hort Sin. 2004;31(5):698.
LinkOut - more resources
Full Text Sources
