Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
- PMID: 37426619
- PMCID: PMC10329278
- DOI: 10.1515/biol-2022-0637
Virtual high-throughput screening: Potential inhibitors targeting aminopeptidase N (CD13) and PIKfyve for SARS-CoV-2
Abstract
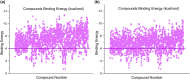
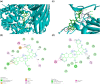
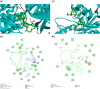
Since the outbreak of the novel coronavirus nearly 3 years ago, the world's public health has been under constant threat. At the same time, people's travel and social interaction have also been greatly affected. The study focused on the potential host targets of SARS-CoV-2, CD13, and PIKfyve, which may be involved in viral infection and the viral/cell membrane fusion stage of SARS-CoV-2 in humans. In this study, electronic virtual high-throughput screening for CD13 and PIKfyve was conducted using Food and Drug Administration-approved compounds in ZINC database. The results showed that dihydroergotamine, Saquinavir, Olysio, Raltegravir, and Ecteinascidin had inhibitory effects on CD13. Dihydroergotamine, Sitagliptin, Olysio, Grazoprevir, and Saquinavir could inhibit PIKfyve. After 50 ns of molecular dynamics simulation, seven compounds showed stability at the active site of the target protein. Hydrogen bonds and van der Waals forces were formed with target proteins. At the same time, the seven compounds showed good binding free energy after binding to the target proteins, providing potential drug candidates for the treatment and prevention of SARS-CoV-2 and SARS-CoV-2 variants.
Keywords: CD13; PIKfyve; SARS-CoV-2; drug repurposing; potential inhibitor; virtual screening.
© 2023 the author(s), published by De Gruyter.
Conflict of interest statement
Conflict of interest: Authors state no conflict of interest.
Figures




Similar articles
-
Computational repurposing approach for targeting the critical spike mutations in B.1.617.2 (delta), AY.1 (delta plus) and C.37 (lambda) SARS-CoV-2 variants using exhaustive structure-based virtual screening, molecular dynamic simulations and MM-PBSA methods.Comput Biol Med. 2022 Aug;147:105709. doi: 10.1016/j.compbiomed.2022.105709. Epub 2022 Jun 7. Comput Biol Med. 2022. PMID: 35728285 Free PMC article.
-
Virtual high throughput screening: Potential inhibitors for SARS-CoV-2 PLPRO and 3CLPRO proteases.Eur J Pharmacol. 2021 Jun 15;901:174082. doi: 10.1016/j.ejphar.2021.174082. Epub 2021 Apr 3. Eur J Pharmacol. 2021. PMID: 33823185 Free PMC article.
-
In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.J Infect Public Health. 2020 Sep;13(9):1210-1223. doi: 10.1016/j.jiph.2020.06.016. Epub 2020 Jun 16. J Infect Public Health. 2020. PMID: 32561274 Free PMC article.
-
Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches.Infect Genet Evol. 2020 Oct;84:104451. doi: 10.1016/j.meegid.2020.104451. Epub 2020 Jul 5. Infect Genet Evol. 2020. PMID: 32640381 Free PMC article.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
Cited by
-
Peptide discovery across the spectrum of neuroinflammation; microglia and astrocyte phenotypical targeting, mediation, and mechanistic understanding.Front Mol Neurosci. 2024 Nov 20;17:1443985. doi: 10.3389/fnmol.2024.1443985. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39634607 Free PMC article. Review.
References
-
- Ahmad W, Gull B, Baby J, Panicker NG, Khader TA, Akhlaq S, et al. Differentially-regulated miRNAs in COVID-19: A systematic review. Rev Med Virol. 2023;e2449. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
