TMEM123 a key player in immune surveillance of colorectal cancer
- PMID: 37426665
- PMCID: PMC10327427
- DOI: 10.3389/fimmu.2023.1194087
TMEM123 a key player in immune surveillance of colorectal cancer
Erratum in
-
Corrigendum: TMEM123 a key player in immune surveillance of colorectal cancer.Front Immunol. 2023 Jul 28;14:1256713. doi: 10.3389/fimmu.2023.1256713. eCollection 2023. Front Immunol. 2023. PMID: 37575239 Free PMC article.
Abstract
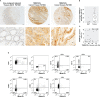
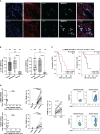
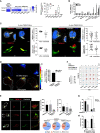
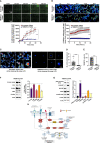
Colorectal cancer (CRC) is a leading cause of cancer-associated death. In the tumor site, the interplay between effector immune cells and cancer cells determines the balance between tumor elimination or outgrowth. We discovered that the protein TMEM123 is over-expressed in tumour-infiltrating CD4 and CD8 T lymphocytes and it contributes to their effector phenotype. The presence of infiltrating TMEM123+ CD8+ T cells is associated with better overall and metastasis-free survival. TMEM123 localizes in the protrusions of infiltrating T cells, it contributes to lymphocyte migration and cytoskeleton organization. TMEM123 silencing modulates the underlying signaling pathways dependent on the cytoskeletal regulator WASP and the Arp2/3 actin nucleation complex, which are required for synaptic force exertion. Using tumoroid-lymphocyte co-culture assays, we found that lymphocytes form clusters through TMEM123, anchoring to cancer cells and contributing to their killing. We propose an active role for TMEM123 in the anti-cancer activity of T cells within tumour microenvironment.
Keywords: TMEM123; cell adhesion; colorectal cancer; cytoskeleton organization; migration; tumor microenvironment; tumor-infiltrating lymphocytes.
Copyright © 2023 Pesce, Cordiglieri, Bombaci, Eppenberger-Castori, Oliveto, Manara, Crosti, Ercan, Coto, Gobbini, Campagnoli, Donnarumma, Martinelli, Bevilacqua, De Camilli, Gruarin, Sarnicola, Cassinotti, Baldari, Viale, Biffo, Abrignani, Terracciano and Grifantini.
Conflict of interest statement
Author Renata Grifantini is currently employed by CheckmAb Srl. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the international immunooncology biomarkers working group: part 1: assessing the host immune response, TILs in invasi. Adv Anat Pathol (2017) 24:235–51. doi: 10.1097/PAP.0000000000000162 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

