Detection of endogenous translesion DNA synthesis in single mammalian cells
- PMID: 37426760
- PMCID: PMC10326377
- DOI: 10.1016/j.crmeth.2023.100501
Detection of endogenous translesion DNA synthesis in single mammalian cells
Abstract
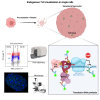
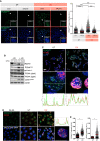
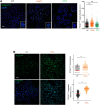
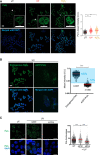
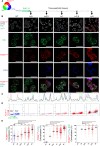
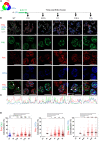
Translesion DNA synthesis (TLS) is an evolutionarily conserved process that cells activate to tolerate DNA damage. TLS facilitates proliferation under DNA damage conditions and is exploited by cancer cells to gain therapy resistance. It has been so far challenging to analyze endogenous TLS factors such as PCNAmUb and TLS DNA polymerases in single mammalian cells due to a lack of suitable detection tools. We have adapted a flow cytometry-based quantitative method allowing detection of endogenous, chromatin-bound TLS factors in single mammalian cells, either untreated or exposed to DNA-damaging agents. This high-throughput procedure is quantitative, accurate, and allows unbiased analysis of TLS factors' recruitment to chromatin, as well as occurrence of DNA lesions with respect to the cell cycle. We also demonstrate detection of endogenous TLS factors by immunofluorescence microscopy and provide insights into TLS dynamics upon DNA replication forks stalled by UV-C-induced DNA damage.
Keywords: CPDs; DNA damage tolerance; PCNAmUb; TLS pols; UV-C; XP-V; chromatin; flow cytometry; oxidative stress; replication stress.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

