Genome of tetraploid sour cherry (Prunus cerasus L.) 'Montmorency' identifies three distinct ancestral Prunus genomes
- PMID: 37426879
- PMCID: PMC10323630
- DOI: 10.1093/hr/uhad097
Genome of tetraploid sour cherry (Prunus cerasus L.) 'Montmorency' identifies three distinct ancestral Prunus genomes
Abstract
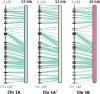
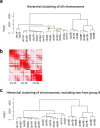
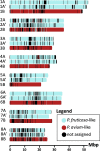
Sour cherry (Prunus cerasus L.) is a valuable fruit crop in the Rosaceae family and a hybrid between progenitors closely related to extant Prunus fruticosa (ground cherry) and Prunus avium (sweet cherry). Here we report a chromosome-scale genome assembly for sour cherry cultivar Montmorency, the predominant cultivar grown in the USA. We also generated a draft assembly of P. fruticosa to use alongside a published P. avium sequence for syntelog-based subgenome assignments for 'Montmorency' and provide compelling evidence P. fruticosa is also an allotetraploid. Using hierarchal k-mer clustering and phylogenomics, we show 'Montmorency' is trigenomic, containing two distinct subgenomes inherited from a P. fruticosa-like ancestor (A and A') and two copies of the same subgenome inherited from a P. avium-like ancestor (BB). The genome composition of 'Montmorency' is AA'BB and little-to-no recombination has occurred between progenitor subgenomes (A/A' and B). In Prunus, two known classes of genes are important to breeding strategies: the self-incompatibility loci (S-alleles), which determine compatible crosses, successful fertilization, and fruit set, and the Dormancy Associated MADS-box genes (DAMs), which strongly affect dormancy transitions and flowering time. The S-alleles and DAMs in 'Montmorency' and P. fruticosa were manually annotated and support subgenome assignments. Lastly, the hybridization event 'Montmorency' is descended from was estimated to have occurred less than 1.61 million years ago, making sour cherry a relatively recent allotetraploid. The 'Montmorency' genome highlights the evolutionary complexity of the genus Prunus and will inform future breeding strategies for sour cherry, comparative genomics in the Rosaceae, and questions regarding neopolyploidy.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
None declared.
Figures

References
-
- Chin S-W, Shaw J, Haberle Ret al. Diversification of almonds, peaches, plums and cherries—molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet Evol. 2014;76:34–48. - PubMed
-
- Hodel RGJ, Zimmer E, Wen J. A phylogenomic approach resolves the backbone of Prunus (Rosaceae) and identifies signals of hybridization and allopolyploidy. Mol Phylogenet Evol. 2021;160:107118. - PubMed
-
- Olden EJ, Nybom N. On the origin of Prunus cerasus L. Hereditas. 1968;59:107118–59.
-
- Beaver JA, Iezzoni AF. Allozyme inheritance in tetraploid sour cherry (Prunus cerasus L.). J Am Soc Hortic. 1993;118:873–7.
LinkOut - more resources
Full Text Sources
Miscellaneous

