Live-Cell Imaging of Drosophila melanogaster Third Instar Larval Brains
- PMID: 37427933
- PMCID: PMC10655794
- DOI: 10.3791/65538
Live-Cell Imaging of Drosophila melanogaster Third Instar Larval Brains
Abstract
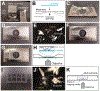
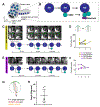
Drosophila neural stem cells (neuroblasts, NBs hereafter) undergo asymmetric divisions, regenerating the self-renewing neuroblast, while also forming a differentiating ganglion mother cell (GMC), which will undergo one additional division to give rise to two neurons or glia. Studies in NBs have uncovered the molecular mechanisms underlying cell polarity, spindle orientation, neural stem cell self-renewal, and differentiation. These asymmetric cell divisions are readily observable via live-cell imaging, making larval NBs ideally suited for investigating the spatiotemporal dynamics of asymmetric cell division in living tissue. When properly dissected and imaged in nutrient-supplemented medium, NBs in explant brains robustly divide for 12-20 h. Previously described methods are technically difficult and may be challenging to those new to the field. Here, a protocol is described for the preparation, dissection, mounting, and imaging of live third-instar larval brain explants using fat body supplements. Potential problems are also discussed, and examples are provided for how this technique can be used.
Figures



References
-
- Homem CCF, Knoblich JA Drosophila neuroblasts: A model for stem cell biology. Development. 139 (23), 4297–4310 (2012). - PubMed
-
- Gallaud E, Pham T, Cabernard C Drosophila melanogaster neuroblasts: A model for asymmetric stem cell divisions. Results and Problems in Cell Differentiation. 61 (1489), 183–210 (2017). - PubMed
-
- Loyer N, Januschke J Where does asymmetry come from? Illustrating principles of polarity and asymmetry establishment in Drosophila neuroblasts. Current Opinion in Cell Biology. 62, 70–77 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
