Breadth and Durability of SARS-CoV-2-Specific T Cell Responses following Long-Term Recovery from COVID-19
- PMID: 37428088
- PMCID: PMC10433967
- DOI: 10.1128/spectrum.02143-23
Breadth and Durability of SARS-CoV-2-Specific T Cell Responses following Long-Term Recovery from COVID-19
Abstract
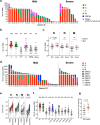
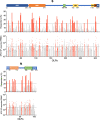
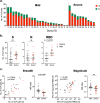
T cell immunity is crucial for long-term immunological memory, but the profile of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific memory T cells in individuals who recovered from COVID-19 (COVID-19-convalescent individuals) is not sufficiently assessed. In this study, the breadth and magnitude of SARS-CoV-2-specific T cell responses were determined in COVID-19-convalescent individuals in Japan. Memory T cells against SARS-CoV-2 were detected in all convalescent individuals, and those with more severe disease exhibited a broader T cell response relative to cases with mild symptoms. Comprehensive screening of T cell responses at the peptide level was conducted for spike (S) and nucleocapsid (N) proteins, and regions frequently targeted by T cells were identified. Multiple regions in S and N proteins were targeted by memory T cells, with median numbers of target regions of 13 and 4, respectively. A maximum of 47 regions were recognized by memory T cells for an individual. These data indicate that SARS-CoV-2-convalescent individuals maintain a substantial breadth of memory T cells for at least several months following infection. Broader SARS-CoV-2-specific CD4+ T cell responses, relative to CD8+ T cell responses, were observed for the S but not the N protein, suggesting that antigen presentation is different between viral proteins. The binding affinity of predicted CD8+ T cell epitopes to HLA class I molecules in these regions was preserved for the Delta variant and at 94 to 96% for SARS-CoV-2 Omicron subvariants, suggesting that the amino acid changes in these variants do not have a major impact on antigen presentation to SARS-CoV-2-specific CD8+ T cells. IMPORTANCE RNA viruses, including SARS-CoV-2, evade host immune responses through mutations. As broader T cell responses against multiple viral proteins could minimize the impact of each single amino acid mutation, the breadth of memory T cells would be one essential parameter for effective protection. In this study, breadth of memory T cells to S and N proteins was assessed in COVID-19-convalescent individuals. While broad T cell responses were induced against both proteins, the ratio of N to S proteins for breadth of T cell responses was significantly higher in milder cases. The breadth of CD4+ and CD8+ T cell responses was also significantly different between S and N proteins, suggesting different contributions of N and S protein-specific T cells for COVID-19 control. Most CD8+ T cell epitopes in the immunodominant regions maintained their HLA binding to SARS-CoV-2 Omicron subvariants. Our study provides insights into understanding the protective efficacy of SARS-CoV-2-specific memory T cells against reinfection.
Keywords: COVID-19; SARS-CoV-2; T cell immunity; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Minimal Crossover between Mutations Associated with Omicron Variant of SARS-CoV-2 and CD8+ T-Cell Epitopes Identified in COVID-19 Convalescent Individuals.mBio. 2022 Apr 26;13(2):e0361721. doi: 10.1128/mbio.03617-21. Epub 2022 Mar 1. mBio. 2022. PMID: 35229637 Free PMC article.
-
Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19.Nat Immunol. 2020 Nov;21(11):1336-1345. doi: 10.1038/s41590-020-0782-6. Epub 2020 Sep 4. Nat Immunol. 2020. PMID: 32887977 Free PMC article.
-
Limited Recognition of Highly Conserved Regions of SARS-CoV-2.Microbiol Spectr. 2022 Feb 23;10(1):e0278021. doi: 10.1128/spectrum.02780-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196796 Free PMC article.
-
Degenerate CD8 Epitopes Mapping to Structurally Constrained Regions of the Spike Protein: A T Cell-Based Way-Out From the SARS-CoV-2 Variants Storm.Front Immunol. 2021 Sep 8;12:730051. doi: 10.3389/fimmu.2021.730051. eCollection 2021. Front Immunol. 2021. PMID: 34566990 Free PMC article. Review.
-
Defending against SARS-CoV-2: The T cell perspective.Front Immunol. 2023 Jan 27;14:1107803. doi: 10.3389/fimmu.2023.1107803. eCollection 2023. Front Immunol. 2023. PMID: 36776863 Free PMC article. Review.
Cited by
-
Long-Term Durability and Variant-Specific Modulation of SARS-CoV-2 Humoral and Cellular Immunity over Two Years.Int J Mol Sci. 2025 Aug 21;26(16):8106. doi: 10.3390/ijms26168106. Int J Mol Sci. 2025. PMID: 40869428 Free PMC article.
-
Post-pandemic memory T cell response to SARS-CoV-2 is durable, broadly targeted, and cross-reactive to the hypermutated BA.2.86 variant.Cell Host Microbe. 2024 Feb 14;32(2):162-169.e3. doi: 10.1016/j.chom.2023.12.003. Epub 2024 Jan 10. Cell Host Microbe. 2024. PMID: 38211583 Free PMC article.
-
Exploring the Pathophysiology of Long COVID: The Central Role of Low-Grade Inflammation and Multisystem Involvement.Int J Mol Sci. 2024 Jun 9;25(12):6389. doi: 10.3390/ijms25126389. Int J Mol Sci. 2024. PMID: 38928096 Free PMC article. Review.
-
Molecular basis of potent antiviral HLA-C-restricted CD8+ T cell response to an immunodominant SARS-CoV-2 nucleocapsid epitope.Nat Commun. 2025 Aug 28;16(1):8062. doi: 10.1038/s41467-025-63288-3. Nat Commun. 2025. PMID: 40877317 Free PMC article.
References
-
- Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S. 2020. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. - DOI - PMC - PubMed
-
- Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, Zhuang Y, Tham CYL, Chia A, Smith GJD, Young B, Kalimuddin S, Low JGH, Lye D, Wang LF, Bertoletti A. 2021. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep 34:108728. doi: 10.1016/j.celrep.2021.108728. - DOI - PMC - PubMed
-
- Tarke A, Potesta M, Varchetta S, Fenoglio D, Iannetta M, Sarmati L, Mele D, Dentone C, Bassetti M, Montesano C, Mondelli MU, Filaci G, Grifoni A, Sette A. 2022. Early and polyantigenic CD4 T cell responses correlate with mild disease in acute COVID-19 donors. Int J Mol Sci 23:7155. doi: 10.3390/ijms23137155. - DOI - PMC - PubMed
-
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, Stralin K, Gorin JB, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, Wullimann DJ, Kammann T, Emgard J, Parrot T, Folkesson E, Karolinska C-SG, Rooyackers O, Eriksson LI, Henter JI, Sonnerborg A, Allander T, Albert J, Nielsen M, Klingstrom J, Gredmark-Russ S, Bjorkstrom NK, Sandberg JK, Price DA, Ljunggren HG, Aleman S, Buggert M, Karolinska COVID-19 Study Group . 2020. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. - DOI - PMC - PubMed
-
- Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, Lopez-Camacho C, Slon-Campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins D, Ker DS, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, de Silva T, Sopp P, Wellington D, Rajapaksa U, Chen YL, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P, ISARIC4C Investigators , et al. 2020. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol 21:1336–1345. doi: 10.1038/s41590-020-0782-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

