Deep learning model for predicting the survival of patients with primary gastrointestinal lymphoma based on the SEER database and a multicentre external validation cohort
- PMID: 37428248
- PMCID: PMC11796771
- DOI: 10.1007/s00432-023-05123-0
Deep learning model for predicting the survival of patients with primary gastrointestinal lymphoma based on the SEER database and a multicentre external validation cohort
Abstract
Purpose: Due to the rarity of primary gastrointestinal lymphoma (PGIL), the prognostic factors and optimal management of PGIL have not been clearly defined. We aimed to establish prognostic models using a deep learning algorithm for survival prediction.
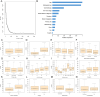
Methods: We collected 11,168 PGIL patients from the Surveillance, Epidemiology, and End Results (SEER) database to form the training and test cohorts. At the same time, we collected 82 PGIL patients from three medical centres to form the external validation cohort. We constructed a Cox proportional hazards (CoxPH) model, random survival forest (RSF) model, and neural multitask logistic regression (DeepSurv) model to predict PGIL patients' overall survival (OS).
Results: The 1-, 3-, 5-, and 10-year OS rates of PGIL patients in the SEER database were 77.1%, 69.4%, 63.7%, and 50.3%, respectively. The RSF model based on all variables showed that the top three most important variables for predicting OS were age, histological type, and chemotherapy. The independent risk factors for PGIL patient prognosis included sex, age, race, primary site, Ann Arbor stage, histological type, symptom, radiotherapy, and chemotherapy, according to the Lasso regression analysis. Using these factors, we built the CoxPH and DeepSurv models. The DeepSurv model's C-index values were 0.760 in the training cohort, 0.742 in the test cohort, and 0.707 in the external validation cohort, which demonstrated that the DeepSurv model performed better compared to the RSF model (0.728) and the CoxPH model (0.724). The DeepSurv model accurately predicted 1-, 3-, 5- and 10-year OS. Both calibration curves and decision curve analysis curves demonstrated the superior performance of the DeepSurv model. We developed the DeepSurv model as an online web calculator for survival prediction, which can be accessed at http://124.222.228.112:8501/ .
Conclusions: This DeepSurv model with external validation is superior to previous studies in predicting short-term and long-term survival and can help us make better-individualized decisions for PGIL patients.
Keywords: Deep learning; DeepSurv; Machine learning; Survival analysis; primary gastrointestinal lymphoma.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures






Similar articles
-
Comparison of Random Survival Forest Based-Overall Survival With Deep Learning and Cox Proportional Hazard Models in HER-2-Positive HR-Negative Breast Cancer.Cancer Rep (Hoboken). 2025 Jul;8(7):e70262. doi: 10.1002/cnr2.70262. Cancer Rep (Hoboken). 2025. PMID: 40624807 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Competing risk and random survival forest models for predicting survival in post-resection elderly stage I-III colorectal cancer patients.Sci Rep. 2025 Jul 7;15(1):24269. doi: 10.1038/s41598-025-05824-1. Sci Rep. 2025. PMID: 40624131 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Optimisation of chemotherapy and radiotherapy for untreated Hodgkin lymphoma patients with respect to second malignant neoplasms, overall and progression-free survival: individual participant data analysis.Cochrane Database Syst Rev. 2017 Sep 13;9(9):CD008814. doi: 10.1002/14651858.CD008814.pub2. Cochrane Database Syst Rev. 2017. PMID: 28901021 Free PMC article.
Cited by
-
Machine learning in image-based outcome prediction after radiotherapy: A review.J Appl Clin Med Phys. 2025 Jan;26(1):e14559. doi: 10.1002/acm2.14559. Epub 2024 Nov 18. J Appl Clin Med Phys. 2025. PMID: 39556691 Free PMC article. Review.
-
Primary duodenal T/histiocyte-rich large B-cell lymphoma complicated with obstructive jaundice: A case report and review of literature.World J Gastrointest Surg. 2025 Jan 27;17(1):99758. doi: 10.4240/wjgs.v17.i1.99758. World J Gastrointest Surg. 2025. PMID: 39872781 Free PMC article.
References
-
- Alvarez-Lesmes J, Chapman JR, Cassidy D, Zhou Y, Garcia-Buitrago M, Montgomery EA, Lossos IS, Sussman D, Poveda J (2021) Gastrointestinal tract lymphomas. Arch Pathol Lab Med 145(12):1585–1596. 10.5858/arpa.2020-0661-RA - PubMed
-
- Cheung MC, Housri N, Ogilvie MP, Sola JE, Koniaris LG (2009) Surgery does not adversely affect survival in primary gastrointestinal lymphoma. J Surg Oncol 100(1):59–64. 10.1002/jso.21298 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

