Large-deviations of disease spreading dynamics with vaccination
- PMID: 37428751
- PMCID: PMC10332629
- DOI: 10.1371/journal.pone.0287932
Large-deviations of disease spreading dynamics with vaccination
Abstract
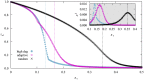
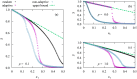
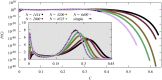
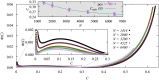
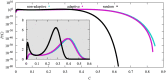
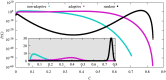
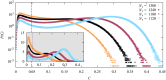
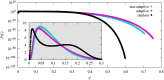
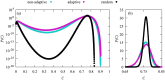
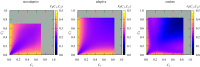
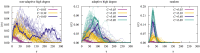
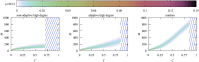
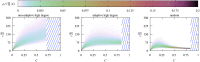
We numerically simulated the spread of disease for a Susceptible-Infected-Recovered (SIR) model on contact networks drawn from a small-world ensemble. We investigated the impact of two types of vaccination strategies, namely random vaccination and high-degree heuristics, on the probability density function (pdf) of the cumulative number C of infected people over a large range of its support. To obtain the pdf even in the range of probabilities as small as 10-80, we applied a large-deviation approach, in particular the 1/t Wang-Landau algorithm. To study the size-dependence of the pdfs within the framework of large-deviation theory, we analyzed the empirical rate function. To find out how typical as well as extreme mild or extreme severe infection courses arise, we investigated the structures of the time series conditioned to the observed values of C.
Copyright: © 2023 Feld, Hartmann. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

