Effects of heterozygous deletion of autism-related gene Cullin-3 in mice
- PMID: 37428799
- PMCID: PMC10332626
- DOI: 10.1371/journal.pone.0283299
Effects of heterozygous deletion of autism-related gene Cullin-3 in mice
Abstract
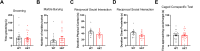
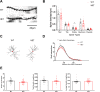
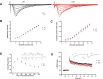
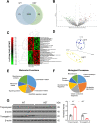
Autism Spectrum Disorder (ASD) is a developmental disorder in which children display repetitive behavior, restricted range of interests, and atypical social interaction and communication. CUL3, coding for a Cullin family scaffold protein mediating assembly of ubiquitin ligase complexes through BTB domain substrate-recruiting adaptors, has been identified as a high-risk gene for autism. Although complete knockout of Cul3 results in embryonic lethality, Cul3 heterozygous mice have reduced CUL3 protein, demonstrate comparable body weight, and display minimal behavioral differences including decreased spatial object recognition memory. In measures of reciprocal social interaction, Cul3 heterozygous mice behaved similarly to their wild-type littermates. In area CA1 of hippocampus, reduction of Cul3 significantly increased mEPSC frequency but not amplitude nor baseline evoked synaptic transmission or paired-pulse ratio. Sholl and spine analysis data suggest there is a small yet significant difference in CA1 pyramidal neuron dendritic branching and stubby spine density. Unbiased proteomic analysis of Cul3 heterozygous brain tissue revealed dysregulation of various cytoskeletal organization proteins, among others. Overall, our results suggest that Cul3 heterozygous deletion impairs spatial object recognition memory, alters cytoskeletal organization proteins, but does not cause major hippocampal neuronal morphology, functional, or behavioral abnormalities in adult global Cul3 heterozygous mice.
Copyright: © 2023 Xia et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared no competing interests except: CMP has been funded in the past by Novartis and reimbursed for travel to speak once each at Dainippon Sumitomo Pharma Co., Pfizer, Roche, Astra-Zeneca, and Psychogenics. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures




References
-
- American Psychiatric Association, American Psychiatric Association. Task Force on DSM-IV. Diagnostic and statistical manual of mental disorders : DSM-IV- TR. 4th ed. Washington, DC: American Psychiatric Association; 2000. xxxvii, 943 p. p.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

