Sequential mutations in exponentially growing populations
- PMID: 37428805
- PMCID: PMC10359018
- DOI: 10.1371/journal.pcbi.1011289
Sequential mutations in exponentially growing populations
Abstract
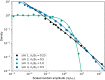
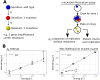
Stochastic models of sequential mutation acquisition are widely used to quantify cancer and bacterial evolution. Across manifold scenarios, recurrent research questions are: how many cells are there with n alterations, and how long will it take for these cells to appear. For exponentially growing populations, these questions have been tackled only in special cases so far. Here, within a multitype branching process framework, we consider a general mutational path where mutations may be advantageous, neutral or deleterious. In the biologically relevant limiting regimes of large times and small mutation rates, we derive probability distributions for the number, and arrival time, of cells with n mutations. Surprisingly, the two quantities respectively follow Mittag-Leffler and logistic distributions regardless of n or the mutations' selective effects. Our results provide a rapid method to assess how altering the fundamental division, death, and mutation rates impacts the arrival time, and number, of mutant cells. We highlight consequences for mutation rate inference in fluctuation assays.
Copyright: © 2023 Nicholson et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
No competing interests to declare.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

