NOS inhibition reverses TLR2-induced chondrocyte dysfunction and attenuates age-related osteoarthritis
- PMID: 37428931
- PMCID: PMC10629581
- DOI: 10.1073/pnas.2207993120
NOS inhibition reverses TLR2-induced chondrocyte dysfunction and attenuates age-related osteoarthritis
Abstract
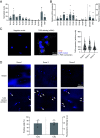
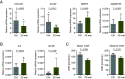
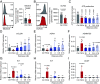
Osteoarthritis (OA) is a joint disease featuring cartilage breakdown and chronic pain. Although age and joint trauma are prominently associated with OA occurrence, the trigger and signaling pathways propagating their pathogenic aspects are ill defined. Following long-term catabolic activity and traumatic cartilage breakdown, debris accumulates and can trigger Toll-like receptors (TLRs). Here we show that TLR2 stimulation suppressed the expression of matrix proteins and induced an inflammatory phenotype in human chondrocytes. Further, TLR2 stimulation impaired chondrocyte mitochondrial function, resulting in severely reduced adenosine triphosphate (ATP) production. RNA-sequencing analysis revealed that TLR2 stimulation upregulated nitric oxide synthase 2 (NOS2) expression and downregulated mitochondria function-associated genes. NOS inhibition partially restored the expression of these genes, and rescued mitochondrial function and ATP production. Correspondingly, Nos2-/- mice were protected from age-related OA development. Taken together, the TLR2-NOS axis promotes human chondrocyte dysfunction and murine OA development, and targeted interventions may provide therapeutic and preventive approaches in OA.
Keywords: Toll-like receptors; cartilage-anabolic and catabolic activities; chondrocytes; nitric oxide synthase; osteoarthritis.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Safiri S., et al. , Global, regional and national burden of osteoarthritis 1990–2017: A systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis. 79, 819–828 (2020). - PubMed
-
- Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J. P., Fahmi H., Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33–42 (2011). - PubMed
-
- Chevalier X., et al. , Intraarticular injection of anakinra in osteoarthritis of the knee: A multicenter, randomized, double-blind, placebo-controlled study. Arthritis Rheum. 61, 344–352 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

