Activity-based protein profiling in microbes and the gut microbiome
- PMID: 37429085
- PMCID: PMC10527501
- DOI: 10.1016/j.cbpa.2023.102351
Activity-based protein profiling in microbes and the gut microbiome
Abstract
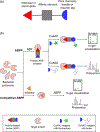
Activity-based protein profiling (ABPP) is a powerful chemical approach for probing protein function and enzymatic activity in complex biological systems. This strategy typically utilizes activity-based probes that are designed to bind a specific protein, amino acid residue, or protein family and form a covalent bond through a reactivity-based warhead. Subsequent analysis by mass spectrometry-based proteomic platforms that involve either click chemistry or affinity-based labeling to enrich for the tagged proteins enables identification of protein function and enzymatic activity. ABPP has facilitated elucidation of biological processes in bacteria, discovery of new antibiotics, and characterization of host-microbe interactions within physiological contexts. This review will focus on recent advances and applications of ABPP in bacteria and complex microbial communities.
Keywords: Activity-based probes; Bacteria; Chemoproteomics; Click chemistry; Gut microbiome.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures

Similar articles
-
[Advances in applications of activity-based chemical probes in the characterization of amino acid reactivities].Se Pu. 2023 Jan;41(1):14-23. doi: 10.3724/SP.J.1123.2022.05013. Se Pu. 2023. PMID: 36633073 Free PMC article. Review. Chinese.
-
Activity-based protein profiling in bacteria: Applications for identification of therapeutic targets and characterization of microbial communities.Curr Opin Chem Biol. 2020 Feb;54:45-53. doi: 10.1016/j.cbpa.2019.10.007. Epub 2019 Dec 10. Curr Opin Chem Biol. 2020. PMID: 31835131 Free PMC article. Review.
-
Activity-Based Protein Profiling-Enabling Multimodal Functional Studies of Microbial Communities.Curr Top Microbiol Immunol. 2019;420:1-21. doi: 10.1007/82_2018_128. Curr Top Microbiol Immunol. 2019. PMID: 30406866 Free PMC article. Review.
-
Application of activity-based protein profiling to study enzyme function in adipocytes.Methods Enzymol. 2014;538:151-69. doi: 10.1016/B978-0-12-800280-3.00009-8. Methods Enzymol. 2014. PMID: 24529438 Free PMC article.
-
Activity- and reactivity-based proteomics: Recent technological advances and applications in drug discovery.Curr Opin Chem Biol. 2021 Feb;60:20-29. doi: 10.1016/j.cbpa.2020.06.011. Epub 2020 Aug 5. Curr Opin Chem Biol. 2021. PMID: 32768892 Review.
Cited by
-
Emerging gut microbial glycoside hydrolase inhibitors.RSC Chem Biol. 2025 Jun 11;6(8):1233-1251. doi: 10.1039/d5cb00050e. eCollection 2025 Jul 30. RSC Chem Biol. 2025. PMID: 40534733 Free PMC article. Review.
-
Activity-based metaproteomics driven discovery and enzymological characterization of potential α-galactosidases in the mouse gut microbiome.Commun Chem. 2024 Aug 16;7(1):184. doi: 10.1038/s42004-024-01273-5. Commun Chem. 2024. PMID: 39152233 Free PMC article.
-
Discovering microbiota functions via chemical probe incorporation for targeted sequencing.Curr Opin Chem Biol. 2025 Feb;84:102551. doi: 10.1016/j.cbpa.2024.102551. Epub 2024 Nov 30. Curr Opin Chem Biol. 2025. PMID: 39615426 Review.
-
The microbiologist's guide to metaproteomics.Imeta. 2025 May 6;4(3):e70031. doi: 10.1002/imt2.70031. eCollection 2025 Jun. Imeta. 2025. PMID: 40469504 Free PMC article. Review.
References
-
- Niphakis MJ, Cravatt BF, Enzyme inhibitor discovery by activity-based protein profiling, Annu. Rev. Biochem 83 (2014) 341–377. - PubMed
-
- Karaj E, Sindi SH, Viranga Tillekeratne LM, Photoaffinity labeling and bioorthogonal ligation: two critical tools for designing “Fish Hooks” to scout for target proteins, Bioorg. Med. Chem 62 (2022) 116721. - PubMed
-
- West AV, Woo CM, Photoaffinity labeling chemistries used to map biomolecular interactions, Isr. J. Chem 63 (2023) e202200081.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

