A Cellular Ground Truth to Develop MRI Signatures in Glioma Models by Correlative Light Sheet Microscopy and Atlas-Based Coregistration
- PMID: 37429718
- PMCID: PMC10376935
- DOI: 10.1523/JNEUROSCI.1470-22.2023
A Cellular Ground Truth to Develop MRI Signatures in Glioma Models by Correlative Light Sheet Microscopy and Atlas-Based Coregistration
Abstract
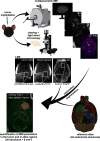
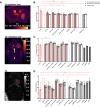
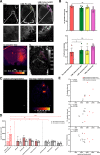
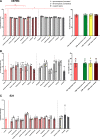
Glioblastoma is the most common malignant primary brain tumor with poor overall survival. Magnetic resonance imaging (MRI) is the main imaging modality for glioblastoma but has inherent shortcomings. The molecular and cellular basis of MR signals is incompletely understood. We established a ground truth-based image analysis platform to coregister MRI and light sheet microscopy (LSM) data to each other and to an anatomic reference atlas for quantification of 20 predefined anatomic subregions. Our pipeline also includes a segmentation and quantification approach for single myeloid cells in entire LSM datasets. This method was applied to three preclinical glioma models in male and female mice (GL261, U87MG, and S24), which exhibit different key features of the human glioma. Multiparametric MR data including T2-weighted sequences, diffusion tensor imaging, T2 and T2* relaxometry were acquired. Following tissue clearing, LSM focused on the analysis of tumor cell density, microvasculature, and innate immune cell infiltration. Correlated analysis revealed differences in quantitative MRI metrics between the tumor-bearing and the contralateral hemisphere. LSM identified tumor subregions that differed in their MRI characteristics, indicating tumor heterogeneity. Interestingly, MRI signatures, defined as unique combinations of different MRI parameters, differed greatly between the models. The direct correlation of MRI and LSM allows an in-depth characterization of preclinical glioma and can be used to decipher the structural, cellular, and, likely, molecular basis of tumoral MRI biomarkers. Our approach may be applied in other preclinical brain tumor or neurologic disease models, and the derived MRI signatures could ultimately inform image interpretation in a clinical setting.SIGNIFICANCE STATEMENT We established a histologic ground truth-based approach for MR image analyses and tested this method in three preclinical glioma models exhibiting different features of glioblastoma. Coregistration of light sheet microscopy to MRI allowed for an evaluation of quantitative MRI data in histologically distinct tumor subregions. Coregistration to a mouse brain atlas enabled a regional comparison of MRI parameters with a histologically informed interpretation of the results. Our approach is transferable to other preclinical models of brain tumors and further neurologic disorders. The method can be used to decipher the structural, cellular, and molecular basis of MRI signal characteristics. Ultimately, information derived from such analyses could strengthen the neuroradiological evaluation of glioblastoma as they enhance the interpretation of MRI data.
Keywords: diffusion tensor imaging; glioma; light sheet microscopy; magnetic resonance imaging; relaxometry; tissue clearing.
Copyright © 2023 the authors.
Figures


Similar articles
-
Assessment of Tumor Cell Invasion and Radiotherapy Response in Experimental Glioma by Magnetic Resonance Elastography.J Magn Reson Imaging. 2025 Mar;61(3):1203-1218. doi: 10.1002/jmri.29567. Epub 2024 Aug 23. J Magn Reson Imaging. 2025. PMID: 39177509 Free PMC article.
-
Imaging signatures of meningioma and low-grade glioma: a diffusion tensor, magnetization transfer and quantitative longitudinal relaxation time MRI study.Magn Reson Imaging. 2016 May;34(4):596-602. doi: 10.1016/j.mri.2015.12.006. Epub 2015 Dec 17. Magn Reson Imaging. 2016. PMID: 26708035 Free PMC article.
-
Characterization of active and infiltrative tumorous subregions from normal tissue in brain gliomas using multiparametric MRI.J Magn Reson Imaging. 2018 Oct;48(4):938-950. doi: 10.1002/jmri.25963. Epub 2018 Feb 7. J Magn Reson Imaging. 2018. PMID: 29412496 Free PMC article.
-
Magnetic resonance relaxometry in quantitative imaging of brain gliomas: A literature review.Neuroradiol J. 2024 Jun;37(3):267-275. doi: 10.1177/19714009231173100. Epub 2023 May 3. Neuroradiol J. 2024. PMID: 37133228 Free PMC article. Review.
-
Susceptibility-Weighted Imaging of Glioma: Update on Current Imaging Status and Future Directions.J Neuroimaging. 2016 Jul;26(4):383-90. doi: 10.1111/jon.12360. Epub 2016 May 26. J Neuroimaging. 2016. PMID: 27227542 Review.
Cited by
-
Three-photon microscopy: an emerging technique for deep intravital brain imaging.Nat Rev Neurosci. 2025 Sep;26(9):521-537. doi: 10.1038/s41583-025-00937-y. Epub 2025 Jun 20. Nat Rev Neurosci. 2025. PMID: 40542182 Review.
-
Revealing the MRI-Contrast in Optically Cleared Brains.Adv Sci (Weinh). 2024 Jun;11(22):e2400316. doi: 10.1002/advs.202400316. Epub 2024 Apr 22. Adv Sci (Weinh). 2024. PMID: 38647385 Free PMC article.
-
Tumor biomechanics as a novel imaging biomarker to assess response to immunotherapy in a murine glioma model.Sci Rep. 2024 Jul 6;14(1):15613. doi: 10.1038/s41598-024-66519-7. Sci Rep. 2024. PMID: 38971907 Free PMC article.
-
Assessment of Tumor Cell Invasion and Radiotherapy Response in Experimental Glioma by Magnetic Resonance Elastography.J Magn Reson Imaging. 2025 Mar;61(3):1203-1218. doi: 10.1002/jmri.29567. Epub 2024 Aug 23. J Magn Reson Imaging. 2025. PMID: 39177509 Free PMC article.
References
-
- Akbari H, Macyszyn L, Da X, Bilello M, Wolf RL, Martinez-Lage M, Biros G, Alonso-Basanta M, O'Rourke DM, Davatzikos C (2016) Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery 78:572–580. 10.1227/NEU.0000000000001202 - DOI - PMC - PubMed
-
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A (2019) ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16:1226–1232. 10.1038/s41592-019-0582-9 - DOI - PubMed
-
- Bobholz SA, Lowman AK, Barrington A, Brehler M, McGarry S, Cochran EJ, Connelly J, Mueller WM, Agarwal M, O'Neill D, Nencka AS, Banerjee A, LaViolette PS (2020) Radiomic features of multiparametric MRI present stable associations with analogous histological features in patients with brain cancer. Tomography 6:160–169. 10.18383/j.tom.2019.00029 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
