Transmission of SARS-CoV-2 in free-ranging white-tailed deer in the United States
- PMID: 37429851
- PMCID: PMC10333304
- DOI: 10.1038/s41467-023-39782-x
Transmission of SARS-CoV-2 in free-ranging white-tailed deer in the United States
Abstract
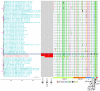
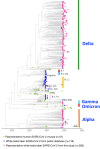
SARS-CoV-2 is a zoonotic virus with documented bi-directional transmission between people and animals. Transmission of SARS-CoV-2 from humans to free-ranging white-tailed deer (Odocoileus virginianus) poses a unique public health risk due to the potential for reservoir establishment where variants may persist and evolve. We collected 8,830 respiratory samples from free-ranging white-tailed deer across Washington, D.C. and 26 states in the United States between November 2021 and April 2022. We obtained 391 sequences and identified 34 Pango lineages including the Alpha, Gamma, Delta, and Omicron variants. Evolutionary analyses showed these white-tailed deer viruses originated from at least 109 independent spillovers from humans, which resulted in 39 cases of subsequent local deer-to-deer transmission and three cases of potential spillover from white-tailed deer back to humans. Viruses repeatedly adapted to white-tailed deer with recurring amino acid substitutions across spike and other proteins. Overall, our findings suggest that multiple SARS-CoV-2 lineages were introduced, became enzootic, and co-circulated in white-tailed deer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- WHO. Tracking SARS-CoV-2 Variants (WHO, 2021)
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

