Lysine methylation promotes NFAT5 activation and determines temozolomide efficacy in glioblastoma
- PMID: 37429858
- PMCID: PMC10333326
- DOI: 10.1038/s41467-023-39845-z
Lysine methylation promotes NFAT5 activation and determines temozolomide efficacy in glioblastoma
Abstract
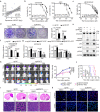
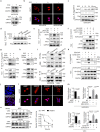
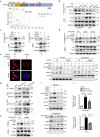
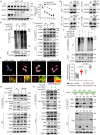
Temozolomide (TMZ) therapy offers minimal clinical benefits in patients with glioblastoma multiforme (GBM) with high EGFR activity, underscoring the need for effective combination therapy. Here, we show that tonicity-responsive enhancer binding protein (NFAT5) lysine methylation, is a determinant of TMZ response. Mechanistically, EGFR activation induces phosphorylated EZH2 (Ser21) binding and triggers NFAT5 methylation at K668. Methylation prevents NFAT5 cytoplasm interaction with E3 ligase TRAF6, thus blocks NFAT5 lysosomal degradation and cytosol localization restriction, which was mediated by TRAF6 induced K63-linked ubiquitination, resulting in NFAT5 protein stabilization, nuclear accumulation and activation. Methylated NFAT5 leads to the upregulation of MGMT, a transcriptional target of NFAT5, which is responsible for unfavorable TMZ response. Inhibition of NFAT5 K668 methylation improved TMZ efficacy in orthotopic xenografts and patient-derived xenografts (PDX) models. Notably, NFAT5 K668 methylation levels are elevated in TMZ-refractory specimens and confer poor prognosis. Our findings suggest targeting NFAT5 methylation is a promising therapeutic strategy to improve TMZ response in tumors with EGFR activation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

