ARIH1 activates STING-mediated T-cell activation and sensitizes tumors to immune checkpoint blockade
- PMID: 37429863
- PMCID: PMC10333294
- DOI: 10.1038/s41467-023-39920-5
ARIH1 activates STING-mediated T-cell activation and sensitizes tumors to immune checkpoint blockade
Abstract
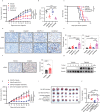
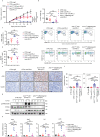
Despite advances in cancer treatment, immune checkpoint blockade (ICB) only achieves complete response in some patients, illustrating the need to identify resistance mechanisms. Using an ICB-insensitive tumor model, here we discover cisplatin enhances the anti-tumor effect of PD-L1 blockade and upregulates the expression of Ariadne RBR E3 ubiquitin-protein ligase 1 (ARIH1) in tumors. Arih1 overexpression promotes cytotoxic T cell infiltration, inhibits tumor growth, and potentiates PD-L1 blockade. ARIH1 mediates ubiquitination and degradation of DNA-PKcs to trigger activation of the STING pathway, which is blocked by the phospho-mimetic mutant T68E/S213D of cGAS protein. Using a high-throughput drug screen, we further identify that ACY738, less cytotoxic than cisplatin, effectively upregulates ARIH1 and activates STING signaling, sensitizing tumors to PD-L1 blockade. Our findings delineate a mechanism that tumors mediate ICB resistance through the loss of ARIH1 and ARIH1-DNA-PKcs-STING signaling and indicate that activating ARIH1 is an effective strategy to improve the efficacy of cancer immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

