ALK peptide vaccination restores the immunogenicity of ALK-rearranged non-small cell lung cancer
- PMID: 37430060
- PMCID: PMC12229965
- DOI: 10.1038/s43018-023-00591-2
ALK peptide vaccination restores the immunogenicity of ALK-rearranged non-small cell lung cancer
Abstract
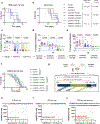
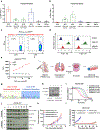
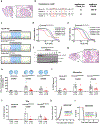
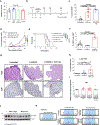
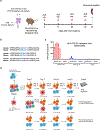
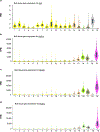
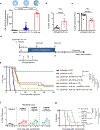
Anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancer (NSCLC) is treated with ALK tyrosine kinase inhibitors (TKIs), but the lack of activity of immune checkpoint inhibitors (ICIs) is poorly understood. Here, we identified immunogenic ALK peptides to show that ICIs induced rejection of ALK+ tumors in the flank but not in the lung. A single-peptide vaccination restored priming of ALK-specific CD8+ T cells, eradicated lung tumors in combination with ALK TKIs and prevented metastatic dissemination of tumors to the brain. The poor response of ALK+ NSCLC to ICIs was due to ineffective CD8+ T cell priming against ALK antigens and is circumvented through specific vaccination. Finally, we identified human ALK peptides displayed by HLA-A*02:01 and HLA-B*07:02 molecules. These peptides were immunogenic in HLA-transgenic mice and were recognized by CD8+ T cells from individuals with NSCLC, paving the way for the development of a clinical vaccine to treat ALK+ NSCLC.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Conflicts of Interest:
R.C. and R.B.B. have filed IP related to human ALK vaccine sequences. R.C. and D.J.I. hold stock options in Elicio Therapeutics. DJI is a consultant to Elicio Therapeutics. R.B.B is a current employee of Moderna Inc. C.J.W. receives research funding from Pharmacyclics and holds equity in BioNTech Inc. D.B.K. is a scientific advisor for Immunitrack and Breakbio. DBK owns equity in Affimed N.V., Agenus, Armata Pharmaceuticals, Breakbio, BioMarin Pharmaceutical, Celldex Therapeutics, Editas Medicine, Gilead Sciences, Immunitybio, ImmunoGen, IMV, Lexicon Pharmaceuticals, Neoleukin Therapeutics. BeiGene, a Chinese biotech company, supported unrelated SARS COV-2 research at TIGL.
The remaining authors declare no competing interests.
Figures







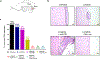

Comment in
-
Peptide vaccine turns up the heat in ALK-positive cancers.Nat Rev Drug Discov. 2023 Sep;22(9):696. doi: 10.1038/d41573-023-00123-8. Nat Rev Drug Discov. 2023. PMID: 37524810 No abstract available.
Similar articles
-
Targeted therapy for advanced anaplastic lymphoma kinase (<I>ALK</I>)-rearranged non-small cell lung cancer.Cochrane Database Syst Rev. 2022 Jan 7;1(1):CD013453. doi: 10.1002/14651858.CD013453.pub2. Cochrane Database Syst Rev. 2022. PMID: 34994987 Free PMC article.
-
Systematic review and network meta-analysis of lorlatinib with comparison to other anaplastic lymphoma kinase (ALK) tyrosine kinase inhibitors (TKIs) as first-line treatment for ALK-positive advanced non-smallcell lung cancer (NSCLC).Lung Cancer. 2024 Nov;197:107968. doi: 10.1016/j.lungcan.2024.107968. Epub 2024 Sep 29. Lung Cancer. 2024. PMID: 39368244
-
Comparative efficacy and safety of first-line treatments for advanced non-small cell lung cancer with ALK-rearranged: a meta-analysis of clinical trials.BMC Cancer. 2021 Nov 26;21(1):1278. doi: 10.1186/s12885-021-08977-0. BMC Cancer. 2021. PMID: 34836510 Free PMC article.
-
Comparative safety of anaplastic lymphoma kinase tyrosine kinase inhibitors in advanced anaplastic lymphoma kinase-mutated non-small cell lung cancer: Systematic review and network meta-analysis.Lung Cancer. 2023 Oct;184:107319. doi: 10.1016/j.lungcan.2023.107319. Epub 2023 Aug 7. Lung Cancer. 2023. PMID: 37597303
-
Effect of ALK-inhibitors in the treatment of non-small cell lung cancer: a systematic review and meta-analysis.Eur Rev Med Pharmacol Sci. 2017 Aug;21(15):3496-3503. Eur Rev Med Pharmacol Sci. 2017. PMID: 28829490
Cited by
-
Development and Clinical Applications of Therapeutic Cancer Vaccines with Individualized and Shared Neoantigens.Vaccines (Basel). 2024 Jun 27;12(7):717. doi: 10.3390/vaccines12070717. Vaccines (Basel). 2024. PMID: 39066355 Free PMC article. Review.
-
Peptide vaccine turns up the heat in ALK-positive cancers.Nat Rev Drug Discov. 2023 Sep;22(9):696. doi: 10.1038/d41573-023-00123-8. Nat Rev Drug Discov. 2023. PMID: 37524810 No abstract available.
-
Highlighting recent achievements to advance more effective cancer immunotherapy.J Exp Clin Cancer Res. 2025 Feb 18;44(1):57. doi: 10.1186/s13046-025-03316-8. J Exp Clin Cancer Res. 2025. PMID: 39966867 Free PMC article.
-
Anti-ALK autoantibodies in patients with ALK-positive Non-Small Cell Lung Cancer (NSCLC): A monocentric experience.J Liq Biopsy. 2024 Jul 30;6:100164. doi: 10.1016/j.jlb.2024.100164. eCollection 2024 Dec. J Liq Biopsy. 2024. PMID: 40027306 Free PMC article.
-
Cancer vaccines: from an immunology perspective.Immunother Adv. 2023 Dec 21;4(1):ltad030. doi: 10.1093/immadv/ltad030. eCollection 2024. Immunother Adv. 2023. PMID: 38223410 Free PMC article. Review.
References
-
- Shaw AT et al. Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): a randomised, controlled, open-label, phase 3 trial. The Lancet. Oncology 18, 874–886, doi: 10.1016/S1470-2045(17)30339-X (2017). - DOI - PubMed
Methods-Only References
-
- Reynisson B, Alvarez B, Paul S, Peters B & Nielsen M NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic acids research 48, W449–W454, doi: 10.1093/nar/gkaa379 (2020). - DOI - PMC - PubMed
-
- Schultze JL et al. Follicular lymphomas can be induced to present alloantigen efficiently: a conceptual model to improve their tumor immunogenicity. Proceedings of the National Academy of Sciences of the United States of America 92, 8200–8204, doi: 10.1073/pnas.92.18.8200 (1995). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

