Whole-body cellular mapping in mouse using standard IgG antibodies
- PMID: 37430076
- PMCID: PMC11021200
- DOI: 10.1038/s41587-023-01846-0
Whole-body cellular mapping in mouse using standard IgG antibodies
Abstract
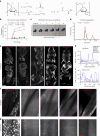
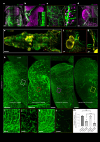
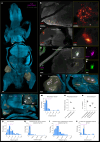
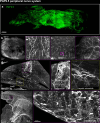
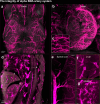
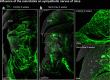
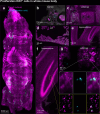
Whole-body imaging techniques play a vital role in exploring the interplay of physiological systems in maintaining health and driving disease. We introduce wildDISCO, a new approach for whole-body immunolabeling, optical clearing and imaging in mice, circumventing the need for transgenic reporter animals or nanobody labeling and so overcoming existing technical limitations. We identified heptakis(2,6-di-O-methyl)-β-cyclodextrin as a potent enhancer of cholesterol extraction and membrane permeabilization, enabling deep, homogeneous penetration of standard antibodies without aggregation. WildDISCO facilitates imaging of peripheral nervous systems, lymphatic vessels and immune cells in whole mice at cellular resolution by labeling diverse endogenous proteins. Additionally, we examined rare proliferating cells and the effects of biological perturbations, as demonstrated in germ-free mice. We applied wildDISCO to map tertiary lymphoid structures in the context of breast cancer, considering both primary tumor and metastases throughout the mouse body. An atlas of high-resolution images showcasing mouse nervous, lymphatic and vascular systems is accessible at http://discotechnologies.org/wildDISCO/atlas/index.php .
© 2023. The Author(s).
Conflict of interest statement
A.E., H.M. and J.L. are also employees of Deep Piction. A.E., H.M. and J.L. filed an intellectual property on technologies described here. The remaining authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

