RNA polymerase II dynamics shape enhancer-promoter interactions
- PMID: 37430091
- PMCID: PMC10714922
- DOI: 10.1038/s41588-023-01442-7
RNA polymerase II dynamics shape enhancer-promoter interactions
Abstract
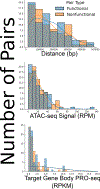
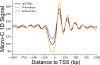
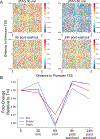
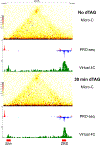
How enhancers control target gene expression over long genomic distances remains an important unsolved problem. Here we investigated enhancer-promoter communication by integrating data from nucleosome-resolution genomic contact maps, nascent transcription and perturbations affecting either RNA polymerase II (Pol II) dynamics or the activity of thousands of candidate enhancers. Integration of new Micro-C experiments with published CRISPRi data demonstrated that enhancers spend more time in close proximity to their target promoters in functional enhancer-promoter pairs compared to nonfunctional pairs, which can be attributed in part to factors unrelated to genomic position. Manipulation of the transcription cycle demonstrated a key role for Pol II in enhancer-promoter interactions. Notably, promoter-proximal paused Pol II itself partially stabilized interactions. We propose an updated model in which elements of transcriptional dynamics shape the duration or frequency of interactions to facilitate enhancer-promoter communication.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Ethics declarations
Competing interests
The authors declare no competing interests.
Figures










Comment in
-
RNA Pol II enters the ring of cohesin-mediated loop extrusion.Nat Genet. 2023 Aug;55(8):1256-1258. doi: 10.1038/s41588-023-01463-2. Nat Genet. 2023. PMID: 37452199 No abstract available.
References
-
- Levine M & Tjian R Transcription regulation and animal diversity. Nature 424, 147–151 (2003). - PubMed
-
- Banerji J, Rusconi S & Schaffner W Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981). - PubMed
-
- Benoist C & Chambon P In vivo sequence requirements of the SV40 early promoter region. Nature 290, 304–310 (1981). - PubMed
-
- Choi OR & Engel JD Developmental regulation of beta-globin gene switching. Cell 55, 17–26 (1988). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

