A novel interface between the N-terminal and coiled-coil domain of STAT1 functions in an auto-inhibitory manner
- PMID: 37430250
- PMCID: PMC10331961
- DOI: 10.1186/s12964-023-01124-1
A novel interface between the N-terminal and coiled-coil domain of STAT1 functions in an auto-inhibitory manner
Abstract
Background: STAT1 is an intracellular signaling molecule that is crucially involved in the regulation of the innate immune system by activation of defense mechanisms against microbial pathogens. Phosphorylation-dependent activation of the STAT1 transcription factor is associated with a conversion from an antiparallel to parallel dimer configuration, which after nuclear import binds to DNA. However, not much is known about the specific intermolecular interactions that stabilize unphosphorylated, antiparallel STAT1 complexes prior to activation.
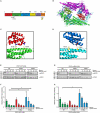
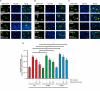
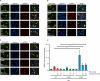
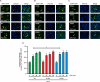
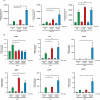
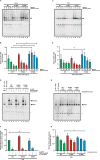
Results: In this study, we identified a previously unknown interdimeric interaction site, which is involved in the termination of STAT1 signaling. Introduction of the glutamic acid-to-alanine point mutation E169A in the coiled-coil domain (CCD) by site-directed mutagenesis led to increased tyrosine phosphorylation as well as accelerated and prolonged nuclear accumulation in transiently transfected cells. In addition, DNA-binding affinity and transcriptional activity were strongly enhanced in the substitution mutant compared to the wild-type (WT) protein. Furthermore, we have demonstrated that the E169 residue in the CCD mediates the release of the dimer from the DNA in an auto-inhibitory manner.
Conclusion: Based on these findings, we propose a novel mechanism for the inactivation of the STAT1 signaling pathway, assigning the interface with the glutamic acid residue 169 in the CCD a crucial role in this process. Video Abstract.
Keywords: Cytokine response; Gene expression; Interferon signaling; STAT signaling; Signal transduction.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Identification of a distinct subset of disease-associated gain-of-function missense mutations in the STAT1 coiled-coil domain as system mutants.Mol Immunol. 2019 Oct;114:30-40. doi: 10.1016/j.molimm.2019.07.008. Epub 2019 Jul 20. Mol Immunol. 2019. PMID: 31336247
-
An inhibitory effect on the nuclear accumulation of phospho-STAT1 by its unphosphorylated form.Cell Commun Signal. 2022 Mar 31;20(1):42. doi: 10.1186/s12964-022-00841-3. Cell Commun Signal. 2022. PMID: 35361236 Free PMC article.
-
A conserved motif in the linker domain of STAT1 transcription factor is required for both recognition and release from high-affinity DNA-binding sites.PLoS One. 2014 May 21;9(5):e97633. doi: 10.1371/journal.pone.0097633. eCollection 2014. PLoS One. 2014. PMID: 24847715 Free PMC article.
-
Clinically relevant dimer interface mutants of STAT1 transcription factor exhibit differential gene expression.PLoS One. 2013 Jul 26;8(7):e69903. doi: 10.1371/journal.pone.0069903. Print 2013. PLoS One. 2013. PMID: 23922848 Free PMC article.
-
Alanine-scanning mutagenesis of human signal transducer and activator of transcription 1 to estimate loss- or gain-of-function variants.J Allergy Clin Immunol. 2017 Jul;140(1):232-241. doi: 10.1016/j.jaci.2016.09.035. Epub 2016 Dec 20. J Allergy Clin Immunol. 2017. PMID: 28011069 Free PMC article.
Cited by
-
Up-regulated Lnc BTU promotes the production of duck plague virus DNA polymerase and inhibits the activation of JAK-STAT pathway to facilitate duck plague virus replication.Poult Sci. 2024 Dec;103(12):104238. doi: 10.1016/j.psj.2024.104238. Epub 2024 Sep 2. Poult Sci. 2024. PMID: 39383668 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

