Whole-genome transcriptome and DNA methylation dynamics of pre-implantation embryos reveal progression of embryonic genome activation in buffaloes
- PMID: 37430306
- PMCID: PMC10334608
- DOI: 10.1186/s40104-023-00894-5
Whole-genome transcriptome and DNA methylation dynamics of pre-implantation embryos reveal progression of embryonic genome activation in buffaloes
Abstract
Background: During mammalian pre-implantation embryonic development (PED), the process of maternal-to-zygote transition (MZT) is well orchestrated by epigenetic modification and gene sequential expression, and it is related to the embryonic genome activation (EGA). During MZT, the embryos are sensitive to the environment and easy to arrest at this stage in vitro. However, the timing and regulation mechanism of EGA in buffaloes remain obscure.
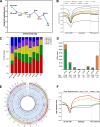
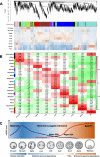
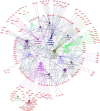
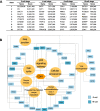
Results: Buffalo pre-implantation embryos were subjected to trace cell based RNA-seq and whole-genome bisulfite sequencing (WGBS) to draw landscapes of transcription and DNA-methylation. Four typical developmental steps were classified during buffalo PED. Buffalo major EGA was identified at the 16-cell stage by the comprehensive analysis of gene expression and DNA methylation dynamics. By weighted gene co-expression network analysis, stage-specific modules were identified during buffalo maternal-to-zygotic transition, and key signaling pathways and biological process events were further revealed. Programmed and continuous activation of these pathways was necessary for success of buffalo EGA. In addition, the hub gene, CDK1, was identified to play a critical role in buffalo EGA.
Conclusions: Our study provides a landscape of transcription and DNA methylation in buffalo PED and reveals deeply the molecular mechanism of the buffalo EGA and genetic programming during buffalo MZT. It will lay a foundation for improving the in vitro development of buffalo embryos.
Keywords: Buffalo; DNA methylome; Embryonic genome activation; Maternal-to-zygote transition; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Embryonic Genome Activation (EGA) Occurred at 1-Cell Stage of Embryonic Development in the Mud Crab, Scylla paramamosain, Revealed by RNA-Seq.Mar Biotechnol (NY). 2024 Dec;26(6):1246-1259. doi: 10.1007/s10126-024-10369-x. Epub 2024 Sep 9. Mar Biotechnol (NY). 2024. PMID: 39249630
-
Embryonic genome activation events in buffalo (Bubalus bubalis) preimplantation embryos.Mol Reprod Dev. 2012 May;79(5):321-8. doi: 10.1002/mrd.22027. Epub 2012 Mar 29. Mol Reprod Dev. 2012. PMID: 22461405
-
Genome activation in bovine embryos: review of the literature and new insights from RNA sequencing experiments.Anim Reprod Sci. 2014 Sep;149(1-2):46-58. doi: 10.1016/j.anireprosci.2014.05.016. Epub 2014 Jun 6. Anim Reprod Sci. 2014. PMID: 24975847 Review.
-
Maternal transcription profiles at different stages for the development of early embryo in buffalo.Reprod Domest Anim. 2020 Apr;55(4):503-514. doi: 10.1111/rda.13644. Epub 2020 Feb 12. Reprod Domest Anim. 2020. PMID: 31971628
-
Epigenetic reprogramming during the maternal-to-zygotic transition.MedComm (2020). 2023 Aug 2;4(4):e331. doi: 10.1002/mco2.331. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37547174 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

