Exploring breast tissue microbial composition and the association with breast cancer risk factors
- PMID: 37430354
- PMCID: PMC10332106
- DOI: 10.1186/s13058-023-01677-6
Exploring breast tissue microbial composition and the association with breast cancer risk factors
Abstract
Background: Microbial dysbiosis has emerged as an important element in the development and progression of various cancers, including breast cancer. However, the microbial composition of the breast from healthy individuals, even relative to risk of developing breast cancer, remains unclear. Here, we performed a comprehensive analysis of the microbiota of the normal breast tissue, which was analyzed in relation to the microbial composition of the tumor and adjacent normal tissue.
Methods: The study cohorts included 403 cancer-free women (who donated normal breast tissue cores) and 76 breast cancer patients (who donated tumor and/or adjacent normal tissue samples). Microbiome profiling was obtained by sequencing the nine hypervariable regions of the 16S rRNA gene (V1V2, V2V3, V3V4, V4V5, V5V7, and V7V9). Transcriptome analysis was also performed on 190 normal breast tissue samples. Breast cancer risk score was assessed using the Tyrer-Cuzick risk model.
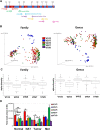
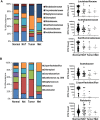
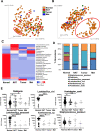
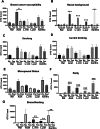
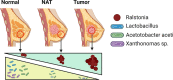
Results: The V1V2 amplicon sequencing resulted more suitable for the analysis of the normal breast microbiome and identified Lactobacillaceae (Firmicutes phylum), Acetobacterraceae, and Xanthomonadaceae (both Proteobacteria phylum) as the most abundant families in the normal breast. However, Ralstonia (Proteobacteria phylum) was more abundant in both breast tumors and histologically normal tissues adjacent to malignant tumors. We also conducted a correlation analysis between the microbiome and known breast cancer risk factors. Abundances of the bacterial taxa Acetotobacter aceti, Lactobacillus vini, Lactobacillus paracasei, and Xanthonomas sp. were associated with age (p < 0.0001), racial background (p < 0.0001), and parity (p < 0.0001). Finally, transcriptome analysis of normal breast tissues showed an enrichment in metabolism- and immune-related genes in the tissues with abundant Acetotobacter aceti, Lactobacillus vini, Lactobacillus paracasei, and Xanthonomas sp., whereas the presence of Ralstonia in the normal tissue was linked to dysregulation of genes involved in the carbohydrate metabolic pathway.
Conclusions: This study defines the microbial features of normal breast tissue, thus providing a basis to understand cancer-related dysbiosis. Moreover, the findings reveal that lifestyle factors can significantly affect the normal breast microbial composition.
Keywords: Acetobacter aceti; Breast cancer risk factors; Lactobacillus; Normal breast; Ralstonia; Xanthomonas sp..
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women.Sci Rep. 2019 Aug 16;9(1):11940. doi: 10.1038/s41598-019-48348-1. Sci Rep. 2019. PMID: 31420578 Free PMC article.
-
Microbiome composition indicate dysbiosis and lower richness in tumor breast tissues compared to healthy adjacent paired tissue, within the same women.BMC Cancer. 2022 Jan 3;22(1):30. doi: 10.1186/s12885-021-09074-y. BMC Cancer. 2022. PMID: 34980006 Free PMC article.
-
Composition and Functional Potential of the Human Mammary Microbiota Prior to and Following Breast Tumor Diagnosis.mSystems. 2022 Jun 28;7(3):e0148921. doi: 10.1128/msystems.01489-21. Epub 2022 Jun 1. mSystems. 2022. PMID: 35642922 Free PMC article.
-
The power of small changes: Comprehensive analyses of microbial dysbiosis in breast cancer.Biochim Biophys Acta Rev Cancer. 2019 Apr;1871(2):392-405. doi: 10.1016/j.bbcan.2019.04.001. Epub 2019 Apr 11. Biochim Biophys Acta Rev Cancer. 2019. PMID: 30981803 Free PMC article. Review.
-
A Systematic Review and Meta-Analysis of 16S rRNA and Cancer Microbiome Atlas Datasets to Characterize Microbiota Signatures in Normal Breast, Mastitis, and Breast Cancer.Microorganisms. 2025 Feb 19;13(2):467. doi: 10.3390/microorganisms13020467. Microorganisms. 2025. PMID: 40005832 Free PMC article. Review.
Cited by
-
Microbiome-Stealth Regulator of Breast Homeostasis and Cancer Metastasis.Cancers (Basel). 2024 Aug 31;16(17):3040. doi: 10.3390/cancers16173040. Cancers (Basel). 2024. PMID: 39272898 Free PMC article. Review.
-
Emerging Role of Gut Microbiota in Breast Cancer Development and Its Implications in Treatment.Metabolites. 2024 Dec 5;14(12):683. doi: 10.3390/metabo14120683. Metabolites. 2024. PMID: 39728464 Free PMC article. Review.
-
Anticancer Potential of Prebiotics: Targeting Estrogen Receptors and PI3K/AKT/mTOR in Breast Cancer.Biomedicines. 2025 Apr 18;13(4):990. doi: 10.3390/biomedicines13040990. Biomedicines. 2025. PMID: 40299687 Free PMC article. Review.
-
Temporal and Spatial Dynamics of Tumor-Host Microbiota in Breast Cancer Progression.Microorganisms. 2025 Jul 10;13(7):1632. doi: 10.3390/microorganisms13071632. Microorganisms. 2025. PMID: 40732141 Free PMC article.
-
Microbial Therapy and Breast Cancer Management: Exploring Mechanisms, Clinical Efficacy, and Integration within the One Health Approach.Int J Mol Sci. 2024 Jan 16;25(2):1110. doi: 10.3390/ijms25021110. Int J Mol Sci. 2024. PMID: 38256183 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

