Structural insights into NMDA receptor pharmacology
- PMID: 37431773
- PMCID: PMC10586783
- DOI: 10.1042/BST20230122
Structural insights into NMDA receptor pharmacology
Abstract
N-methyl-d-aspartate receptors (NMDARs) comprise a subfamily of ionotropic glutamate receptors that form heterotetrameric ligand-gated ion channels and play fundamental roles in neuronal processes such as synaptic signaling and plasticity. Given their critical roles in brain function and their therapeutic importance, enormous research efforts have been devoted to elucidating the structure and function of these receptors and developing novel therapeutics. Recent studies have resolved the structures of NMDARs in multiple functional states, and have revealed the detailed gating mechanism, which was found to be distinct from that of other ionotropic glutamate receptors. This review provides a brief overview of the recent progress in understanding the structures of NMDARs and the mechanisms underlying their function, focusing on subtype-specific, ligand-induced conformational dynamics.
Keywords: glutamate receptor; neuropharmacology; structure.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

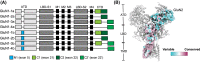
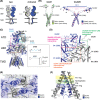

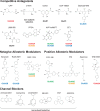
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

