Indolo[2,3- e]benzazocines and indolo[2,3- f]benzazonines and their copper(II) complexes as microtubule destabilizing agents
- PMID: 37431840
- PMCID: PMC10368209
- DOI: 10.1039/d3dt01632c
Indolo[2,3- e]benzazocines and indolo[2,3- f]benzazonines and their copper(II) complexes as microtubule destabilizing agents
Abstract
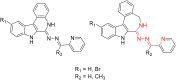
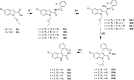
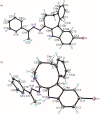
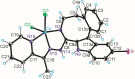
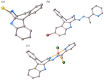
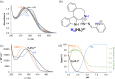
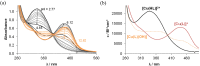
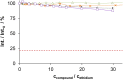
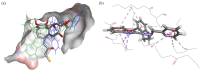
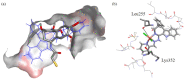
A series of four indolo[2,3-e]benzazocines HL1-HL4 and two indolo[2,3-f]benzazonines HL5 and HL6, as well as their respective copper(II) complexes 1-6, were synthesized and characterized by 1H and 13C NMR spectroscopy, ESI mass spectrometry, single crystal X-ray diffraction (SC-XRD) and combustion analysis (C, H, N). SC-XRD studies of precursors Vd, VIa·0.5MeOH, of ligands HL4 and HL6·DCM, and complexes 2·2DMF, 5·2DMF, 5'·iPrOH·MeOH provided insights into the energetically favored conformations of eight- and nine-membered heterocycles in the four-ring systems. In addition, proton dissociation constants (pKa) of HL1, HL2 and HL5, complexes 1, 2 and 5, overall stability constants (log β) of 1, 2 and 5 in 30% (v/v) DMSO/H2O at 298 K, as well as thermodynamic solubility of HL1-HL6 and 1-6 in aqueous solution at pH 7.4 were determined by UV-vis spectroscopy. All compounds were tested for antiproliferative activity against Colo320, Colo205 and MCF-7 cell lines and showed IC50 values in the low micromolar to sub-micromolar concentration range, while some of them (HL1, HL5 and HL6, 1, 2 and 6) showed remarkable selectivity towards malignant cell lines. Ethidium bromide displacement studies provided evidence that DNA is not the primary target for these drugs. Rather, inhibition of tubulin assembly is likely the underlying mechanism responsible for their antiproliferative activity. Tubulin disassembly experiments showed that HL1 and 1 are effective microtubule destabilizing agents binding to the colchicine site. This was also confirmed by molecular modelling investigations. To the best of our knowledge, complex 1 is the first reported transition metal complex to effectively bind to the tubulin-colchicine pocket.
Conflict of interest statement
The authors declare no competing interest.
Figures
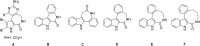




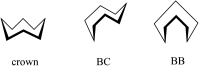

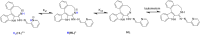


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

