GWLD: an R package for genome-wide linkage disequilibrium analysis
- PMID: 37431944
- PMCID: PMC10468308
- DOI: 10.1093/g3journal/jkad154
GWLD: an R package for genome-wide linkage disequilibrium analysis
Abstract
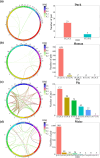
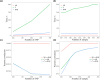
Linkage disequilibrium (LD) analysis is fundamental to the investigation of the genetic architecture of complex traits (e.g. human disease, animal and plant breeding) and population structure and evolution dynamics. However, until now, studies primarily focus on LD status between genetic variants located on the same chromosome. Moreover, genome (re)sequencing produces unprecedented numbers of genetic variants, and fast LD computation becomes a challenge. Here, we have developed GWLD, a parallelized and generalized tool designed for the rapid genome-wide calculation of LD values, including conventional D/D', r2, and (reduced) mutual information (MI and RMI) measures. LD between genetic variants within and across chromosomes can be rapidly computed and visualized in either an R package or a standalone C++ software package. To evaluate the accuracy and speed of LD calculation, we conducted comparisons using 4 real datasets. Interchromosomal LD patterns observed potentially reflect levels of selection intensity across different species. Both versions of GWLD, the R package (https://github.com/Rong-Zh/GWLD/GWLD-R) and the standalone C++ software (https://github.com/Rong-Zh/GWLD/GWLD-C++), are freely available on GitHub.
Keywords: C++; Rcpp package; genome-wide analysis; linkage disequilibrium; mutual information; reduced mutual information.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
