Mrs4 loss of function in fungi during adaptation to the cystic fibrosis lung
- PMID: 37432019
- PMCID: PMC10470810
- DOI: 10.1128/mbio.01171-23
Mrs4 loss of function in fungi during adaptation to the cystic fibrosis lung
Abstract
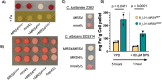
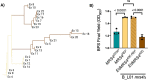
The genetic disease cystic fibrosis (CF) frequently leads to chronic lung infections by bacteria and fungi. We identified three individuals with CF with persistent lung infections dominated by Clavispora (Candida) lusitaniae. Whole-genome sequencing analysis of multiple isolates from each infection found evidence for selection for mutants in the gene MRS4 in all three distinct lung-associated populations. In each population, we found one or two unfixed, non-synonymous mutations in MRS4 relative to the reference allele found in multiple environmental and clinical isolates including the type strain. Genetic and phenotypic analyses found that all evolved alleles led to loss of function (LOF) of Mrs4, a mitochondrial iron transporter. RNA-seq analyses found that Mrs4 variants with decreased activity led to increased expression of genes involved in iron acquisition mechanisms in both low iron and replete iron conditions. Furthermore, surface iron reductase activity and intracellular iron were much higher in strains with Mrs4 LOF variants. Parallel studies found that a subpopulation of a CF-associated Exophiala dermatitidis infection also had a non-synonymous LOF mutation in MRS4. Together, these data suggest that MRS4 mutations may be beneficial during chronic CF lung infections in diverse fungi, perhaps, for the purposes of adaptation to an iron-restricted environment with chronic infections. IMPORTANCE The identification of MRS4 mutations in Clavispora (Candida) lusitaniae and Exophiala dermatitidis in individuals with cystic fibrosis (CF) highlights a possible adaptive mechanism for fungi during chronic CF lung infections. The findings of this study suggest that loss of function of the mitochondrial iron transporter Mrs4 can lead to increased activity of iron acquisition mechanisms, which may be advantageous for fungi in iron-restricted environments during chronic infections. This study provides valuable information for researchers working toward a better understanding of the pathogenesis of chronic lung infections and more effective therapies to treat them.
Keywords: Candida albicans; Clavispora lusitaniae; Exophiala dermatitidis; Mrs4; cystic fibrosis; fungi; iron.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Mrs4 loss of function in fungi during adaptation to the cystic fibrosis lung.bioRxiv [Preprint]. 2023 Apr 6:2023.04.05.535776. doi: 10.1101/2023.04.05.535776. bioRxiv. 2023. Update in: mBio. 2023 Aug 31;14(4):e0117123. doi: 10.1128/mbio.01171-23. PMID: 37066389 Free PMC article. Updated. Preprint.
References
-
- Kim SH, Clark ST, Surendra A, Copeland JK, Wang PW, Ammar R, Collins C, Tullis DE, Nislow C, Hwang DM, Guttman DS, Cowen LE, Hogan DA. 2015. Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor nrg1 as a mechanism of pathogen adaptation. PLoS Pathog 11:e1005308. doi:10.1371/journal.ppat.1005308 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

