Single-strain behavior predicts responses to environmental pH and osmolality in the gut microbiota
- PMID: 37432034
- PMCID: PMC10470613
- DOI: 10.1128/mbio.00753-23
Single-strain behavior predicts responses to environmental pH and osmolality in the gut microbiota
Abstract
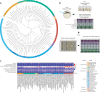
Changes to gut environmental factors such as pH and osmolality due to disease or drugs correlate with major shifts in microbiome composition; however, we currently cannot predict which species can tolerate such changes or how the community will be affected. Here, we assessed the growth of 92 representative human gut bacterial strains spanning 28 families across multiple pH values and osmolalities in vitro. The ability to grow in extreme pH or osmolality conditions correlated with the availability of known stress response genes in many cases, but not all, indicating that novel pathways may participate in protecting against acid or osmotic stresses. Machine learning analysis uncovered genes or subsystems that are predictive of differential tolerance in either acid or osmotic stress. For osmotic stress, we corroborated the increased abundance of these genes in vivo during osmotic perturbation. The growth of specific taxa in limiting conditions in isolation in vitro correlated with survival in complex communities in vitro and in an in vivo mouse model of diet-induced intestinal acidification. Our data show that in vitro stress tolerance results are generalizable and that physical parameters may supersede interspecies interactions in determining the relative abundance of community members. This study provides insight into the ability of the microbiota to respond to common perturbations that may be encountered in the gut and provides a list of genes that correlate with increased ability to survive in these conditions. IMPORTANCE To achieve greater predictability in microbiota studies, it is crucial to consider physical environmental factors such as pH and particle concentration, as they play a pivotal role in influencing bacterial function and survival. For example, pH is significantly altered in various diseases, including cancers, inflammatory bowel disease, as well in the case of over-the-counter drug use. Additionally, conditions like malabsorption can affect particle concentration. In our study, we investigate how changes in environmental pH and osmolality can serve as predictive indicators of bacterial growth and abundance. Our research provides a comprehensive resource for anticipating shifts in microbial composition and gene abundance during complex perturbations. Moreover, our findings underscore the significance of the physical environment as a major driver of bacterial composition. Finally, this work emphasizes the necessity of incorporating physical measurements into animal and clinical studies to better understand the factors influencing shifts in microbiota abundance.
Keywords: acid stress; culturomics; machine learning; microbiota; osmolality; single-strain culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Evaluation of gut microbiota predictive potential associated with phenotypic characteristics to identify multifactorial diseases.Gut Microbes. 2024 Jan-Dec;16(1):2297815. doi: 10.1080/19490976.2023.2297815. Epub 2024 Jan 18. Gut Microbes. 2024. PMID: 38235595 Free PMC article.
-
The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection.Microbiol Spectr. 2015 Jun;3(3). doi: 10.1128/microbiolspec.MBP-0008-2014. Microbiol Spectr. 2015. PMID: 26185088
-
Transient Osmotic Perturbation Causes Long-Term Alteration to the Gut Microbiota.Cell. 2018 Jun 14;173(7):1742-1754.e17. doi: 10.1016/j.cell.2018.05.008. Cell. 2018. PMID: 29906449 Free PMC article.
-
Gut-on-a-Chip for the Analysis of Bacteria-Bacteria Interactions in Gut Microbial Community: What Would Be Needed for Bacterial Co-Culture Study to Explore the Diet-Microbiota Relationship?Nutrients. 2023 Feb 23;15(5):1131. doi: 10.3390/nu15051131. Nutrients. 2023. PMID: 36904133 Free PMC article. Review.
-
Effects of exercise and physical activity on gut microbiota composition and function in older adults: a systematic review.BMC Geriatr. 2023 Jun 12;23(1):364. doi: 10.1186/s12877-023-04066-y. BMC Geriatr. 2023. PMID: 37308839 Free PMC article.
Cited by
-
Composition, Influencing Factors, and Effects on Host Nutrient Metabolism of Fungi in Gastrointestinal Tract of Monogastric Animals.Animals (Basel). 2025 Mar 1;15(5):710. doi: 10.3390/ani15050710. Animals (Basel). 2025. PMID: 40075993 Free PMC article. Review.
-
Proton-pump inhibitors increase C. difficile infection risk by altering pH rather than by affecting the gut microbiome based on a bioreactor model.Gut Microbes. 2025 Dec;17(1):2519697. doi: 10.1080/19490976.2025.2519697. Epub 2025 Jun 16. Gut Microbes. 2025. PMID: 40524314 Free PMC article.
-
Survey of gut microbial biogeography and their functional niche in the grow-finishing swine of ordinary feeding.Front Microbiol. 2025 Mar 7;16:1530553. doi: 10.3389/fmicb.2025.1530553. eCollection 2025. Front Microbiol. 2025. PMID: 40124893 Free PMC article.
-
PUPpy: a primer design pipeline for substrain-level microbial detection and absolute quantification.mSphere. 2024 Jul 30;9(7):e0036024. doi: 10.1128/msphere.00360-24. Epub 2024 Jul 9. mSphere. 2024. PMID: 38980072 Free PMC article.
-
Online monitored characterization of Phocaeicola vulgatus for organic acid production using anaerobic microtiter plate cultivations.Biotechnol Prog. 2025 Mar-Apr;41(2):e3526. doi: 10.1002/btpr.3526. Epub 2024 Dec 20. Biotechnol Prog. 2025. PMID: 39704382 Free PMC article.
References
-
- Strus M, Gosiewski T, Fyderek K, Wedrychowicz A, Kowalska-Duplaga K, Kochan P, Adamski P, Heczko PB. 2009. A role of hydrogen peroxide producing commensal bacteria present in colon of adolescents with inflammatory bowel disease in perpetuation of the inflammatory process. J Physiol Pharmacol 60 Suppl 6:49–54. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

