Studying Membrane Protein-Lipid Specificity through Direct Native Mass Spectrometric Analysis from Tunable Proteoliposomes
- PMID: 37432128
- PMCID: PMC10932607
- DOI: 10.1021/jasms.3c00110
Studying Membrane Protein-Lipid Specificity through Direct Native Mass Spectrometric Analysis from Tunable Proteoliposomes
Abstract
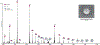
Native mass spectrometry (nMS) has emerged as a key analytical tool to study the organizational states of proteins and their complexes with both endogenous and exogenous ligands. Specifically, for membrane proteins, it provides a key analytical dimension to determine the identity of bound lipids and to decipher their effects on the observed structural assembly. We recently developed an approach to study membrane proteins directly from intact and tunable lipid membranes where both the biophysical properties of the membrane and its lipid compositions can be customized. Extending this, we use our liposome-nMS platform to decipher the lipid specificity of membrane proteins through their multiorganelle trafficking pathways. To demonstrate this, we used VAMP2 and reconstituted it in the endoplasmic reticulum (ER), Golgi, synaptic vesicle (SV), and plasma membrane (PM) mimicking liposomes. By directly studying VAMP2 from these customized liposomes, we show how the same transmembrane protein can bind to different sets of lipids in different organellar-mimicking membranes. Considering that the cellular trafficking pathway of most eukaryotic integral membrane proteins involves residence in multiple organellar membranes, this study highlights how the lipid-specificity of the same integral membrane protein may change depending on the membrane context. Further, leveraging the capability of the platform to study membrane proteins from liposomes with curated biophysical properties, we show how we can disentangle chemical versus biophysical properties, of individual lipids in regulating membrane protein assembly.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
Figures
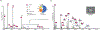


Similar articles
-
Direct determination of oligomeric organization of integral membrane proteins and lipids from intact customizable bilayer.Nat Methods. 2023 Jun;20(6):891-897. doi: 10.1038/s41592-023-01864-5. Epub 2023 Apr 27. Nat Methods. 2023. PMID: 37106230 Free PMC article.
-
Native mass spectrometry of proteoliposomes containing integral and peripheral membrane proteins.Chem Sci. 2023 Nov 21;14(48):14243-14255. doi: 10.1039/d3sc04938h. eCollection 2023 Dec 13. Chem Sci. 2023. PMID: 38098719 Free PMC article.
-
Different regions of synaptic vesicle membrane regulate VAMP2 conformation for the SNARE assembly.Nat Commun. 2020 Mar 24;11(1):1531. doi: 10.1038/s41467-020-15270-4. Nat Commun. 2020. PMID: 32210233 Free PMC article.
-
Fake It 'Till You Make It-The Pursuit of Suitable Membrane Mimetics for Membrane Protein Biophysics.Int J Mol Sci. 2020 Dec 23;22(1):50. doi: 10.3390/ijms22010050. Int J Mol Sci. 2020. PMID: 33374526 Free PMC article. Review.
-
Advances in nanopatterned and nanostructured supported lipid membranes and their applications.Biotechnol Genet Eng Rev. 2010;27:185-216. doi: 10.1080/02648725.2010.10648150. Biotechnol Genet Eng Rev. 2010. PMID: 21415898 Review.
Cited by
-
Structural studies of the human α1 glycine receptor via site-specific chemical cross-linking coupled with mass spectrometry.Biophys Rep (N Y). 2024 Dec 11;4(4):100184. doi: 10.1016/j.bpr.2024.100184. Epub 2024 Oct 10. Biophys Rep (N Y). 2024. PMID: 39393591 Free PMC article.
-
A proteome-wide quantitative platform for nanoscale spatially resolved extraction of membrane proteins into native nanodiscs.Nat Methods. 2025 Feb;22(2):412-421. doi: 10.1038/s41592-024-02517-x. Epub 2024 Nov 28. Nat Methods. 2025. PMID: 39609567 Free PMC article.
-
Structure and function of the human apoptotic scramblase Xkr4.bioRxiv [Preprint]. 2024 Aug 9:2024.08.07.607004. doi: 10.1101/2024.08.07.607004. bioRxiv. 2024. Update in: Nat Commun. 2025 Aug 8;16(1):7317. doi: 10.1038/s41467-025-62739-1. PMID: 39149361 Free PMC article. Updated. Preprint.
-
Native Top-Down Analysis of Membrane Protein Complexes Directly From In Vitro and Native Membranes.Mol Cell Proteomics. 2025 Jul;24(7):100993. doi: 10.1016/j.mcpro.2025.100993. Epub 2025 May 14. Mol Cell Proteomics. 2025. PMID: 40378922 Free PMC article.
-
A proteome-wide quantitative platform for nanoscale spatially resolved extraction of membrane proteins into native nanodiscs.bioRxiv [Preprint]. 2024 Aug 4:2024.02.10.579775. doi: 10.1101/2024.02.10.579775. bioRxiv. 2024. Update in: Nat Methods. 2025 Feb;22(2):412-421. doi: 10.1038/s41592-024-02517-x. PMID: 38405833 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

